Identification of an enzyme that labels lipopolysaccharide on the bacterial surface with ubiquitin marks bacteria for destruction
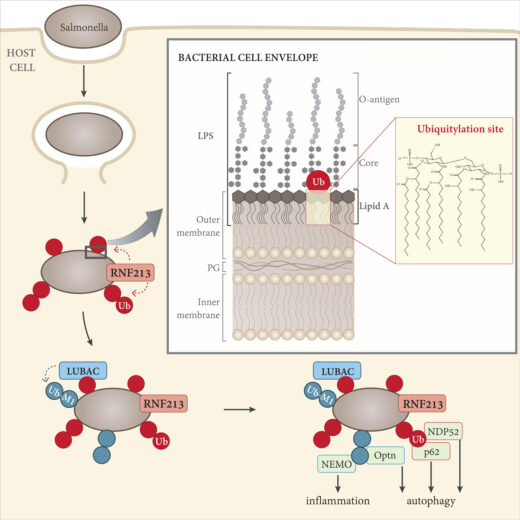
Ubiquitylation is a common post-translational protein modification that affects many aspects of physiology through a variety of mechanisms. Ubiquitylation controls the activity and cellular localisation of proteins, it either enables or prevents protein-protein interactions and it marks proteins for degradation. Ubiquitylation also labels pathogenic bacteria that invade the cytosol of our cells as cargo for anti-bacterial autophagy, a process that is essential to keep our nutrient-rich cytosol sterile. Up until now, it had been understood that ubiquitin is attached exclusively to other proteins, while the identity of the ubiquitylated substrate on invading bacteria had remained unknown. Now, Felix Randow’s group, in our PNAC Division, has shown that, when coating invading Salmonella bacteria, ubiquitin is conjugated directly to the membrane of bacteria, specifically to large non-protein molecules known as lipopolysaccharides (LPS). This is the first observed instance of ubiquitylation of a non-protein target.
Once inside our cells, bacteria are shielded from much of the body’s immune system, so cell-autonomous defences are required to antagonise their survival. One such defence involves capture of invading bacteria inside autophagosomes. Autophagy is activated by the ‘eat-me’ signals galectin 8 and ubiquitin, but the process by which the bacterial ubiquitin coat is created, and the precise targets for ubiquitylation, had remained unknown.
The initial surprising finding that LPS is a substrate of ubiquitylation was made by Emma Werner, a former PhD student in Felix’s group, through the use of a clever genetic approach in Salmonella. Subsequent research led by Gisela Otten, a postdoctoral scientist in the Randow lab, identified RNF213 as the enzyme responsible for LPS ubiquitylation. She used an “old-school” but elegant biochemical approach to purify the enzymatic activity from cell lysates.
Salmonella (red) inside a host cell under attack from RNF213 (green).
The study of RNF213 was complex and technically demanding, as the enzyme is particularly large, being comprised of over 5000 amino acids. In a further twist from the established paradigm, Gisela found that RNF213 catalysis does not rely on any of the domains typically used by eukaryotic ubiquitin ligases. Instead, LPS ubiquitylation depends upon a newly identified domain: the RZ finger. Further work is underway to understand mechanistically how RNF213 deploys its RZ finger to ubiquitylate LPS and possibly other non-canonical substrates.
In addition to discovering a novel type of ubiquitylation, the paper has further significance as mutations in RNF213 are an identified risk factor for the development of Moyamoya disease – a rare disorder characterised by the narrowing of the carotid artery which causes strokes, especially in children. Certain Moyamoya-associated RNF213 alleles are commonly carried, particularly in Asia, but pathological manifestation is relatively uncommon, suggesting that environmental factors could be a critical trigger to the disease. This discovery of how RNF213 responds to and protects against infection, implies a possible role for infection as an important environmental trigger for Moyamoya disease.
As the first example of ubiquitylation of a non-protein substrate, the implications of this study are significant, as it has the potential to expand the scope of ubiquitylation considerably. For example, are there further non-protein substrates of ubiquitylation as well as non-canonical E3 ligases still to be discovered?
The work was funded by UKRI MRC, the Boehringer Ingelheim Trust, and the Wellcome Trust.
Further References
Ubiquitylation of lipopolysaccharide by RNF213 during bacterial infection. Otten, EG., Werner, E., Crespillo Casado, A., Boyle, KB., Dharamdasani, V., Pathe, C., Santhanam, B., Randow, F. Nature
Felix’s group page