Development of an in vitro system shows how the human DNA replication machinery achieves fast and efficient DNA replication
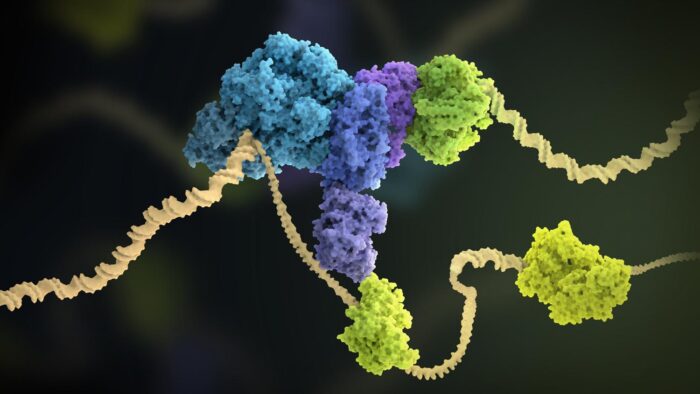
Faithful chromosome replication is essential for the maintenance of genome stability. The complex molecular machinery responsible for this process is the replisome. Joe Yeeles’ group, in the LMB’s PNAC Division, has produced the first biochemical reconstitution of a functional human replisome. The complex was built using 11 purified human replication factors, made up of 43 polypeptides, and is capable of performing fast and efficient DNA replication. The new system allows Joe’s group to gain a better understanding of how these replication factors contribute to human DNA replication.
During DNA replication, the parental double-stranded DNA is unwound by a helicase enzyme to generate single-stranded DNA templates that are then copied by DNA polymerases. Because the two strands of DNA are arranged in an antiparallel configuration and DNA polymerases can only copy DNA in one direction, one strand, called the leading strand, is replicated continuously, while the other strand, termed the lagging strand, is replicated discontinuously in short fragments.
Yasemin Baris, a PhD student in Joe’s group, assembled replisomes on a model forked DNA template, starting with the CMG helicase and then adding the remaining replication proteins. Crucially, Yasemin demonstrated that these replisomes could perform efficient leading and lagging strand DNA replication.
The new system has revealed a number of important insights into fundamental aspects of human DNA replication. The group identified a minimal set of replication factors required to reconstitute replication rates comparable to those measured in cells, and uncovered how these factors contribute to smooth replication fork progression.
The human replisome uses two high-fidelity DNA polymerases, Polε and Polδ. The group showed that Polε is the main polymerase on the leading strand, whereas Polδ functions on the lagging strand. Surprisingly, they found that leading-strand replication by Polε is highly dependent on PCNA, a ring-shaped protein that aids polymerase function, and the CTF18-RFC complex that loads PCNA onto DNA. Conversely, a different PCNA loader complex called RFC was found to be essential for Pol δ function on the lagging strand. These results thus reveal a division of labour for PCNA loading in the human replisome.
Prior to this study, many studies relied on in vitro budding yeast DNA replication systems. However, although much of the replisome is well conserved between yeast and human, budding yeast lacks important proteins and protein domains which are vital to the human DNA replication process. Thus, this new system allows for novel insights specific to the human replisome. One such finding is that, in contrast to the budding yeast replisome, the AND-1 protein directly stimulates leading-strand replication using a region of the protein that is absent from the yeast ortholog.
This breakthrough study will allow researchers to study human replisome structure, replication coupled DNA repair, and recombination pathways to better understand how human-specific factors directly function in these processes. Additionally, dysregulation of the DNA replication process can lead to genome instability, one of the hallmarks of cancer. Replisomes facing exogenous and endogenous stress, or replisome components that have become mutated, have also been identified as a characteristic of cancerous cells. This new in vitro DNA replication system may enable the design of specific inhibitors for disease associated replisome components, pointing towards future possibilities for targeted cancer treatments.
This work was funded by UKRI MRC and The Wellcome Trust.
Further references
Fast and efficient DNA replication with purified human proteins. Baris, Y., Taylor, MRG., Aria, V., Yeeles, JTP. Nature
Joe’s group page
Previous Insight on Research
Regulating the disassembly of the eukaryotic DNA replication machinery
Unravelling the replisome: how a molecular machine overcomes obstacles to DNA replication
How replication of DNA is initiated at origins