Chromosome replication is one of the most crucial cellular processes. How the final step of this process – disassembly of the DNA replication machinery, known as the replisome – is regulated has been revealed for the first time
Faithful chromosome replication is critical for correct cellular function. Mistakes made during chromosome replication can generate mutations or genome rearrangements that cause genomic instability, leading to the development of cancer. Disassembly of the DNA replication machinery, known as the replisome, is the final step of eukaryotic chromosome replication. Exquisite regulation of this process is essential to ensure that only those replisomes which are no longer active are disassembled, and not those which are mid-way through the process of replicating DNA. Premature removal of the active replisomes would have disastrous consequences, causing DNA replication forks to collapse and leading to under-replication of the genome. Research, by Joe Yeeles’ group in the LMB’s PNAC Division, working with Tom Deegan, at the MRC Protein Phosphorylation and Ubiquitination Unit, Dundee (now at the MRC Human Genetics Unit, Edinburgh), reveals for the first time how replication fork DNA itself regulates replisome disassembly.
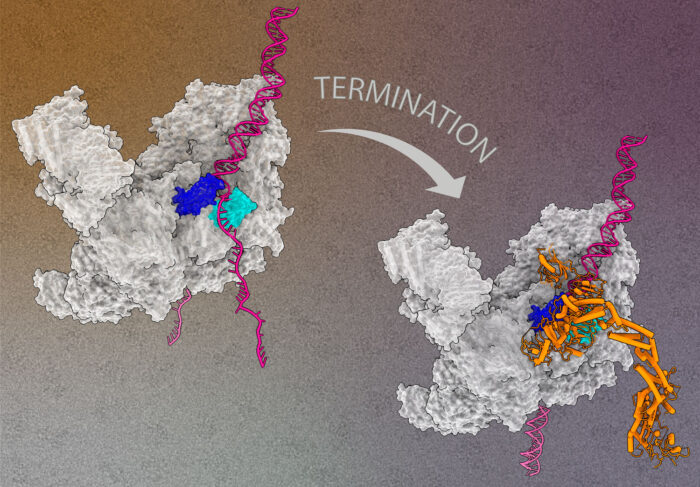
Disassembly of the replisome is controlled by a ubiquitin ligase, either SCFDia2 in yeast or CUL2LRR1 in human. Work by Michael Jenkyn-Bedford, in Joe’s group, determined the structures of yeast replisomes bound to SCFDia2, both on double-strand DNA and in the absence of DNA, using electron cryo-microscopy (cryo-EM). Michael’s structures represent the first bona fide replisome disassembly intermediates to be visualised. Then, using the structures to guide mutagenesis experiments, collaborators were able to identify a previously unknown protein-to-protein interaction site between SCFDia2 and the replisome, showing that this structure was essential for replisome disassembly.
Morgan Jones, also in Joe’s group, then determined the structure of a human replisome containing CUL2LRR1. Although Dia2 and LRR1 are not evolutionarily related proteins, Morgan’s structure showed that both bind to exactly the same part of the replisome, at the zinc finger domains of the Mcm3 and Mcm5 subunits. In a separate recent publication (Jones et al., EMBO J, 2021), Morgan has shown that the so-called excluded strand of the replication fork DNA exits the replisome between these zinc finger domains, thereby occupying exactly the same region of the replisome that both Dia2 and LRR1 bind to.
“Remarkably, although the key protein complex that is involved in replisome disassembly is not shared between yeast and human, the mechanism of regulation we have discovered is conserved between the two species, suggesting that it arose by convergent evolution.” – Joe Yeeles.
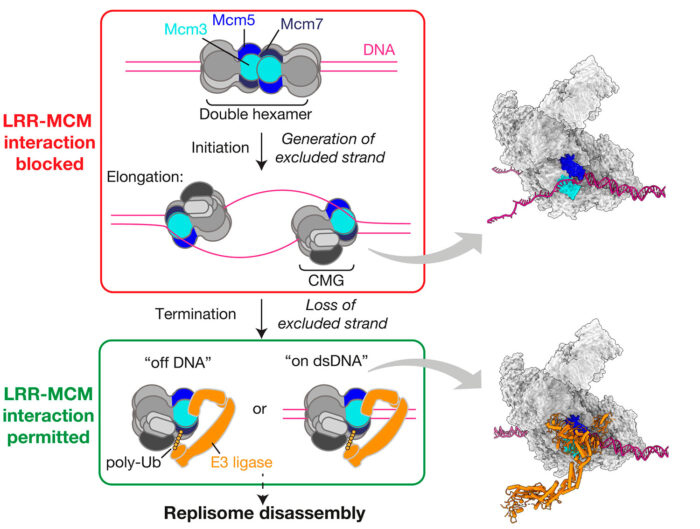
Before this research, scientists were aware that replication fork DNA somehow controls the activity of SCFDia2/CUL2LRR1 to restrict Mcm7 modification and, in doing so, the disassembly of the replisome. However, the mechanism by which this is achieved was entirely unknown. Now, this exciting new work sheds light on how the regulation of replisome disassembly is achieved through a simple and conserved mechanism whereby the SCFDia2/CUL2LRR1 binding site, which is essential for replisome ubiquitination, is sterically blocked. By blocking this site until DNA replication is complete, the excluded DNA strand helps to prevent premature replisome disassembly, protecting the cell from genomic instability. By better understanding the process of DNA replication, there is huge potential that research of this kind will be fundamental in developing specific cancer therapeutics.
This work was funded by the UKRI MRC and the Wellcome Trust.
Further references
A Conserved Mechanism for Regulating Replisome Disassembly in Eukaryotes. Jenkyn-Bedford, M., Jones, M.L., Baris, Y., Labib, K. P. M., Cannone, G., Yeeles, J. T. P. and Deegan, T.D. Nature (2021). https://doi.org/10.1038/s41586-021-04145-3
Joe Yeeles’ group
Tom Deegan’s group (MRC Human Genetics Unit)
Previous Insight on Research
Unravelling the replisome: how a molecular machine overcomes obstacles to DNA replication
How replication of DNA is initiated at origins
How does the replisome respond to DNA damage?