Initiation of DNA synthesis during chromosome replication depends on the actions of the Pol α-primase complex. Joseph Yeeles’ group show how this complex is recruited to, and coordinated by, the DNA replication machinery to begin replication.
Chromosome replication is a crucial cellular process that must be tightly regulated to prevent genome instability. The complex molecular machine responsible for coordinating DNA replication is termed the replisome. During DNA replication, the replisome unwinds double-stranded DNA to generate single-stranded DNA templates for the replicative polymerases to act upon. As the two DNA strands are antiparallel, one strand, termed the leading strand, is replicated continuously, while the second strand, termed the lagging strand, is replicated discontinuously in short fragments.
To initiate DNA replication, the replisome must first recruit the Pol α-primase complex. This complex synthesises short stretches of RNA/DNA called primers on the lagging-strand template which are then extended by the replicative polymerases. Due to the discontinuous nature of lagging-strand DNA replication, Pol α-primase is required to synthesise primers every few hundred nucleotides, which equates to millions of priming events during each cell cycle. A new study by Joseph Yeeles’ group in the LMB’s PNAC Division has revealed the mechanism, conserved across eukaryotes, by which Pol α-primase is recruited to the replisome to prime eukaryotic DNA replication.
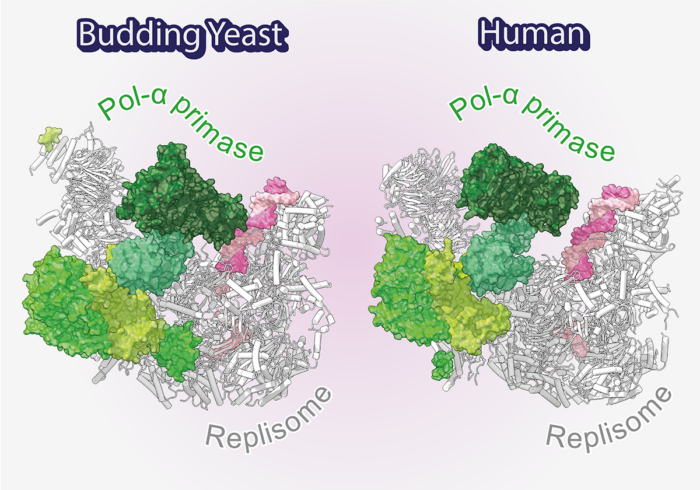
Using electron cryo-microscopy (cryo-EM) Morgan Jones determined the first structures of yeast and human replisomes bound to Pol α-primase. These structures reveal a complex network of interactions that position Pol α-primase on the leading edge of the central replisome component, the CMG replicative helicase, responsible for unwinding double-stranded DNA. These interactions position the primase catalytic subunit directly above the point of DNA strand separation, where it is poised to capture and prime the lagging-strand template. This close proximity to the lagging-, rather than leading-strand template, explains previous work from the Yeeles group showing that, in contrast to lagging-strand priming, leading-strand priming is highly inefficient and therefore lagging-strand primers are used to start leading-strand DNA replication.
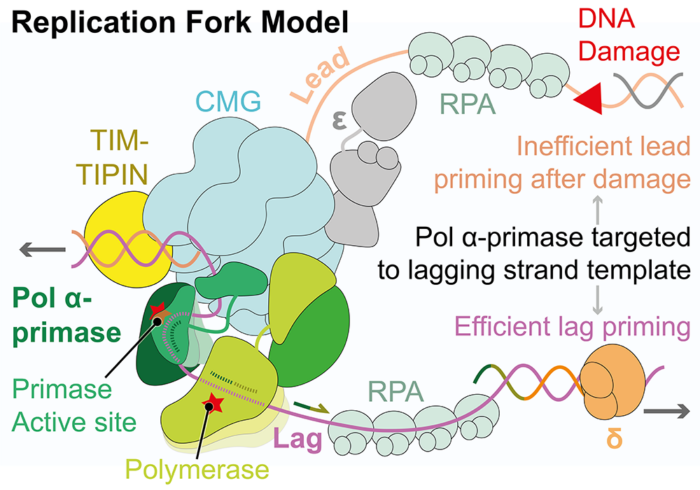
In order to understand how the newly identified interactions between Pol α-primase and the replisome contribute to priming, structure guided mutations were designed to disrupt each site, and their impact assessed using both biochemical and cellular assays. This work revealed a crucial role for the PRIM2/Pri2 subunit of Pol α-primase which forms two conserved interfaces with the replisome that are essential for maintaining normal rates of priming. Before this research, scientists were aware that Pol α-primase is tethered to the replisome via interactions with an additional replisome component. However, considerable evidence indicated that this link was not the only mode of replisome recruitment. Now, this exciting work sheds new light on how Pol α-primase is recruited to the replisome and is able to overcome competition for the lagging strand from the single-strand DNA binding protein RPA.
This study marks a significant breakthrough in the understanding of a fundamental biological process and provides a valuable platform to visualize other essential steps in DNA replication. A detailed understanding of the mechanisms of DNA replication will be crucial in the future design of anti-cancer therapeutics.
This work was funded by UKRI MRC.
Further references
How Pol α-primase is targeted to replisomes to prime eukaryotic DNA replication. Jones, ML., Aria, V., Baris, Y., Yeeles, JTP. Molecular Cell
The Initial Response of a Eukaryotic Replisome to DNA Damage. Taylor MRG, Yeeles JTP. Molecular Cell
Mechanism of bidirectional leading-strand synthesis establishment at eukaryotic DNA replication origins. Aria, V. Yeeles, JTP. Molecular Cell
Previous Insight on Research articles
Functional human replisome reconstituted for the first time
Regulating the disassembly of the eukaryotic DNA replication machinery
Unravelling the replisome: how a molecular machine overcomes obstacles to DNA replication