
Blue Sky Collaboration

AstraZeneca photo by Hufton and Crow
In 2014 a research fund was set up between the MRC Laboratory of Molecular Biology (LMB) and AstraZeneca to support a range of pre-clinical research projects. The aim is to improve our understanding of fundamental biology and disease and encourage innovative scientific thinking by sharing knowledge and technologies.
Since inception, AstraZeneca has contributed approximately £12 million ($20 million), and LMB approximately £6 million ($10 million) and in-kind scientific input, under the collaboration.
Projects involve scientists from the two organisations working side by side, either within the LMB building on the Cambridge Biomedical Campus, or in AstraZeneca research facilities. Projects supported by the fund are not specifically targeted towards drug development but feed into existing research and development activities of the two organisations, with results published in peer reviewed journals.
A Joint Steering Committee (JSC) of LMB and AstraZeneca staff decide which projects will receive support from the Fund.
“This partnership has further strengthened our existing, long-standing relationship with AstraZeneca, allowing scientists to work together, share ideas and expertise”
Jan Löwe, LMB Director
Current Joint Steering Committee Members
- Jan Löwe, LMB Director and Chair
- Simon Bullock, LMB Group Leader
- Marta Zlatic, LMB Group Leader
- Julian Sale, LMB Group Leader
- Susan Critchlow, Executive Director and Head of Oncology Biosciences, AstraZeneca
- Steve Rees, Senior Vice President, Life Sciences, R&D, AstraZeneca
- Maria Groves, Senior Director Oncology R&D, AstraZeneca
Joint Steering Committee Coordinators:
- Chris Phillips, Senior Director, Structural Biology, Discovery Sciences R&D, AstraZeneca
- Sybille Kubis-Waller, LMB, Joint Steering Committee Secretary
Cryo-EM collaboration solves ATM structure
Matching LMB cryo-EM know-how with AstraZeneca reagents and target molecule expertise
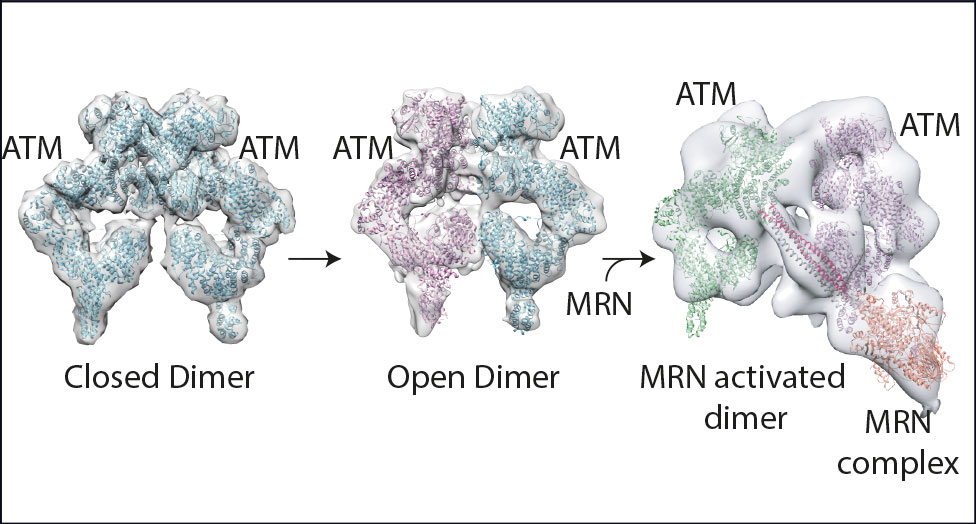
The Blue Sky collaboration between Roger Williams, Group Leader at the LMB, and Chris Phillips, Associate Director and structural biologist with AstraZeneca, resulted in the first three-dimensional structural model of ATM in 2017. ATM (ataxia-telangiectasia mutated) is a key regulator of the DNA damage response (DDR) signalling pathway and target for cancer therapies.
This structural research – with Roger, Chris, and Domagoj Baretić and Anna Howes from the LMB – suggested an activation mechanism for ATM that involves a key transition. This modification from tightly coupled dimers (see graphic) through loosely coupled dimers to a final complex with two molecules of ATM associated with an activated DDR complex at a site of DNA damage was significant. Arka Chakraborty is carrying on structural work on a related complex, ATR/ATRIP.
“Until this paper no three-dimensional structural model was available for ATM and this work has established a framework to interpret past and future ATM mechanistic biology studies. It represents a step change in our understanding of the molecular basis of ATM function.”
Chris Phillips, Associate Director and structural biologist with AstraZeneca
The project benefitted both sides of the collaboration. Hannah Pollard, David Fisher, and Caroline Truman from Chris’s team at AstraZeneca initially cloned, expressed and characterised ATM. “The project got a fast start because we were able to take advantage of the expertise and reagents that AstraZeneca had developed for ATM”, says Roger.
By working with Roger – a world expert in this class of proteins – Chris was able to access Roger’s insight, ideas and experience.
And significantly, this project was AstraZeneca’s first experience with electron cryo-microscopy (cryo-EM), a technique in-part developed at the LMB and for which Richard Henderson was awarded the Chemistry Nobel Prize in 2017. “The LMB in general is the world’s leading EM institute so to have access to this know-how and infrastructure was simply fantastic”, says Chris.
The collaboration enabled both parties to understand the activation mechanism of ATM, but Chris’s team have also used the same methods and reagents in their work aimed at developing inhibitors of these DDR enzymes.
This work proceeded in parallel with cryo-EM for another DDR enzyme by Chris Phillip’s group at AstraZeneca. Chris adds that the collaboration will continue. “We have started work on ATR, a related enzyme in the family, and continue the ATM project. It is a case of watch this space.”

“The collaboration with AstraZeneca has been an amazing experience. At every step of the process we learned from each other and shaped the approaches to our common goal.“
Roger Williams, LMB Group Leader
Publications and press:
Structures of closed and open conformations of dimeric human ATM. Baretic, D., Pollard, H.K., Fisher, D.I., Johnson, C.M., Santhanam, B., Truman, C.M., Kouba, T., Fersht, A.R., Phillips, C., Williams, R.L. Science Advances 2017
AstraZeneca and LMB in Cambridge use one of world’s most advanced microscopes to make breakthrough, Cambridge Independent 2017
A Blue Sky path to new cancer therapy targets
Investigating the link between amoebae feeding regulation and cancer cell growth
Blue Sky funding enabled Rob Kay, now Emeritus Scientist at LMB, and Matthew King, Senior Scientist at AstraZeneca, to investigate an intriguing link between the regulation of a feeding process in lower organisms – macropinocytosis – and its adaptation to promote cancer growth in humans.
Like all living organisms, cancer cells need to feed to survive and grow. Cancer cells capture amino acids and glucose from surrounding medium using specialised transporters in the plasma membrane. These fluids contain another, richer, source of nutrients in the form of proteins and other macromolecules. And to access these, many cancers have adopted an ancient feeding process that they share with amoebae, known as macropinocytotic feeding.
Rob’s research has focused on identifying molecular components of signaling and membrane dynamics in amoebae, including the process of macropinocytosis (see movie). By collaborating with Rob, Matthew was able to tap into his keen understanding of this process at a level of detail sometimes overlooked in disease research. “The investigation of such a concept would normally be thought outside the remit of a pharmaceutical research environment,” says Matthew.
Rob’s group had identified proteins that control macropinocytotic feeding in amoebae. By working together, the team found that two of these, the tumour suppressors NF1 and PTEN, probably had similar functions in mammalian cells. “This has since been confirmed in publications from other laboratories,” says Rob.
The team then sought further parallels between growth factor signalling and macropinocytosis. This research showed that all of the following are required for macropinocytosis in Dictyostelium amoebae – protein kinases Akt and SGK, which act after PIP3 in the signalling cascade, together with their activating kinases, PDK1 and TORC2.
This work further suggests that cytoskeletal proteins that are regulated by the Ras-PIP3-Akt/SGK pathway are also potential targets.

“Growth factor signalling is a major target for cancer therapeutics and macropinocytic feeding is a potential target.”
Rob Kay, LMB Emeritus Scientist
Both parties found collaboration and the flexibility of Blue Sky funding to be beneficial.
“We learned a lot from our collaborator Matt King, who helped design the project and educated us in the ways of cancer cells. When we needed to change direction, the funding was flexible. This allowed us to complete two papers and suggest a very Blue Sky approach to finding new targets for cancer therapy,” says Rob.

“The collaboration gave our research postdoc access to imaging platforms and technology that were not available at either facility alone.”
Matthew King, Senior Scientist at AstraZeneca
Publications
Akt and SGK protein kinases are required for efficient feeding by macropinocytosis. Williams, T.D., Peak-Chew, S.Y., Paschke, P., Kay, R.R. J Cell Sci 2019
Living on soup: macropinocytic feeding in amoebae. Kay, R.R., Williams, T.D., Manton, J.D., Traynor, D., Paschke, P. Int J Dev Biol 2019
How do heart cells tell the time?
Collaborative explorations into how our body clocks work at the cellular level
In 2015 John O’Neill, Group Leader at the LMB, and Peter Newham, Vice President, Global Head of Oncology Safety, AstraZeneca, began an exciting partnership to explore circadian mechanisms in heart cells.
John’s group is interested in how cells operate on a 24-hour rhythm in most aspects of human health and disease. For instance, it has been known for decades that the heart functions most effectively in the afternoon; whereas adverse cardiovascular events, such as sudden cardiac death, occur more frequently in early morning hours. But we don’t know why.
In 2015, John’s group observed that sodium and potassium levels had a circadian, approximately daily, rhythm in many different mammalian cells. “And we were trying to figure out the underlying mechanism and its functional consequences,” says John.
Peter and his colleague Alex Harmer’s scientific interests lay in the physiology of heart cells or cardiomyocytes (see image). “Since disturbances in daily cardiovascular rhythms are associated with increased risk of cardiovascular morbidity, we set out to explore mechanisms for circadian regulation of membrane excitability and contractility in cardiomyocytes. Understanding these processes could greatly assist in considering ‘chrono-pharmacology’, i.e. alignment of drug pharmacodynamics with body clocks,” says Peter.
Image courtesy of Linda Starnes, AstraZeneca R&D
When Peter, Alex and John first met at the 2015 AstraZeneca-LMB Blue Sky meeting, it soon became clear that there was a perfect fit between Peter’s interest in the physiology of heart cells, and John’s biological question.

“Sodium and potassium levels are essential for the electrical activity that allows the heart to beat. So, teaming up with Peter to look at the origin of these ion rhythms in the heart cells and how it affects their daily activity formed the basis of our exciting project together.”
John O’Neill, LMB Group Leader
Working with the right scientists was key to the collaboration’s success. John recruited Alessandra Stangherlin, who had extensive experience with both circadian research and cardiomyocytes. After quickly disproving their initial hypothesis, the project got back on track after invaluable input from Peter and Alex, the use of AstraZeneca resources, some unexpected observations by David Wong – a PhD student in John’s lab, and careful experimental design.
“We finally found that ion rhythms are driven by changes in cytosolic macromolecule concentration, and ultimately dependent on daily rhythms of mTORC signalling and protein synthesis,” says John. mTORC is a protein complex that regulates protein synthesis.
Both John and Peter agree that they were the right scientific partners. Peter adds, “Working with world leaders in dissecting fundamental biological processes has been invigorating and a fantastic learning experience opening up new appreciations of not only circadian biology, but also ingenious ways to unpick the mechanics of cellular biochemistry.

“Our collaboration has been a great opportunity to follow the science, engage in open and collaborative research with the LMB and make truly novel discoveries.”
Peter Newham, Vice President, Global Head of Oncology Safety, AstraZeneca
And the collaboration continues. “This work has led to a further related project in which we’re developing human organoid models that will allow us to understand and predict what time of day different drug treatments will be most effective – in the heart and other tissues,” says John.
Publications and press:
Compensatory ion transport buffers daily protein rhythms to regulate osmotic balance and cellular physiology. Stangherlin, A., Watson, J.L., Wong, D.C.S., Barbiero, S., Zeng, A., Seinkmane, E., Peak Chew, S., Beale, A.D., Hayter, E.A., Guna, A., Inglis, A.J., Putker, M., Bartolami, E., Matile, S., Lequeux, N., Pons, T., Day, J., van Ooijen, G., Voorhees, R.M., Bechtold, D.A., Derivery, E., Edgar, R.S., Newham, P., O’Neill, J.S. Nature Communications 2021
CRYPTOCHROMES promote daily protein homeostasis. Wong, D.C.S., Seinkmane, E., Zeng, A., Stangherlin, A., Rzechorzek, N.M., Beale, A.D., Day, J., Reed, M., Peak-Chew, S.Y., Styles, C.T., Edgar, R.S., Putker, M., O’Neill, J.S. EMBO Journal 2022
Video: How do cellular clocks influence cardiovascular health? astrazeneca 2021
New tools for the nucleic acid chemical toolbox
Capitalising on complimentary expertise and technology know-how
The Blue Sky collaboration between Philipp Holliger, Group Leader at the LMB, and Ralph Minter, until recently Senior Director, Fellow at Astrazeneca (now Head of Research at Alchemab), yielded new tools for the nucleic acid chemical toolbox, specifically for the synthesis, reverse transcription and evolution of chemically modified nucleic acids.
Philipp’s group specialises in the generation of artificial genetic systems and the synthesis and evolution of novel, DNA-like polymers for applications in nanotechnology and material science. Ralph’s expertise lies in antibody discovery, protein evolution and intracellular delivery of macromolecules.

“Ralph and I met at the 2015 AstraZeneca-LMB Blue Sky meeting and initiated discussions on a range of ideas that capitalised on our complimentary expertise and technology know-how.”
Philipp Holliger, LMB Group Leader
Initially, scientists in Philipp’s group focused on the development of technology – the chemical tools – needed to unlock the potential of modified nucleic acids.
The research led by Sebastian Arangundy Franklin together with Benjamin Porebski and Alex Taylor led to the discovery of the first ever polymerase for an uncharged genetic polymer as well as the first ever evolution of an aptamer with an uncharged backbone chemistry. This study showed that genetic function, i.e. heredity and evolution, are possible – in contrast to previous proposals – in the absence of a charged backbone. Similar uncharged genetic polymers are important components of approved anti-sense nucleic acid drugs.
In further work Sebastian Arangundy Franklin, Benjamin Porebski and Nithya Subramanian aided Gillian Houlihan and Alex Taylor in the development and application of a new selection method and the discovery of a range of novel reverse transcriptase enzymes (RTs), including for RTs for xeno-nucleic acid (XNA) chemistries for which previously none existed. Furthermore they showed the versatility of the new method with the discovery of RNA RT enzymes with the highest fidelity ever described.
Development of these new tools was substantially aided by input from Ralph and colleagues: “The team at AstraZeneca provided great technical expertise on selection technology and target selection, as well as overall scientific input”, says Philipp.
Now that the tools are in place, further ambitious collaborative projects utilising modified nucleic acids can be accomplished. Philipp is working on a new Blue Sky-funded project in collaboration with Ben Taylor, Associate Director at AstraZeneca, on developing a truly novel fully in-vitro Cas evolution system. This protein evolution system will allow the teams to take Cas enzyme and screen for mutations that make them work faster, more efficiently and more specifically – properties that are critically needed for research tools and therapeutic applications.

“Success of this project will set a new precedent for how the field develops and hones the activity and specificity of genome engineering enzymes, in terms of speed, simplicity, robustness and tunability to a desired environment, with potential for profound impact on cell manipulation, diagnostics and cell therapies.”
Ben Taylor, Associate Director, AstraZeneca
Publications
A synthetic genetic polymer with an uncharged backbone chemistry based on alkyl phosphonate nucleic acids. Arangundy-Franklin, S., Taylor, A.I., Porebski, B.T., Genna, V., Peak-Chew, S., Vaisman, A., Woodgate, R., Orozco, M., Holliger P. Nature Chem 2019
Discovery and evolution of RNA and XNA reverse transcriptase function andfidelity. Houlihan, G., Arangundy-Franklin, S., Porebski, B.T., Subramanian, N., Taylor, A.I., Holliger, P. Nature Chem 2020
Unlocking the therapeutic potential of Trim-Away
Combining AstraZeneca therapeutic agents with LMB protein depletion technology
Blue Sky funding is enabling LMB and AstraZeneca to explore the therapeutic potential of Trim-Away, a rapid and selective protein depletion technology developed at LMB in 2017.
The collaborative project, led by Leo James, Group Leader at the LMB, James Hunt, Associate Director, Antibody Discovery and Protein Engineering, R&D, AstraZeneca and Mark McAlister, Associate Director, Protein Structure & Biophysics, Discovery Sciences, R&D, AstraZeneca, aims to combine AstraZeneca’s platform technology together with LMB’s novel Trim-Away assay to degrade therapeutically interesting proteins.
Many diseases, such as cancer and neurodegeneration, are caused when cellular protein complexes go wrong. Depletion of these pathogenic or misfunctioning proteins from cells in living organisms can treat, correct or prevent disease. But targeting specific proteins and controlling degradation levels has proven challenging.
In Trim-Away, antibodies are delivered into cells, bind their specific protein target, are recognised by TRIM21, and then destroyed within minutes. VHH are small antibody-derived molecules that can be engineered to bind to specific protein targets within cells which are inaccessible to whole antibodies.
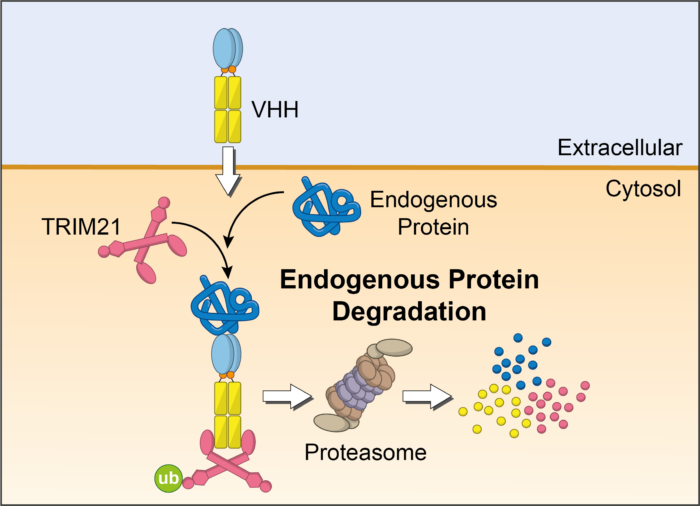
Together researchers in James’s and Leo’s teams investigated whether VHHs could be engineered into chimeric molecules capable of targeting Trim-Away degradation to specific disease-causing proteins in pre-clinical cell models.

“We successfully produced chimeric VHH-based molecules that selectively removed therapeutic protein targets linked to respiratory disease and cancer in cell models.”
Leo James LMB Group Leader
“The targets we selected have proven to be difficult to study using traditional small molecule tools. The opportunity to work together with Leo and Dean to investigate these with TRIM-Away has opened up a new way to study the effects of rapid, targeted protein degradation; an approach that has great therapeutic potential.”
James Hunt, Associate Director, Antibody Discovery and Protein Engineering, R&D, AstraZeneca
In a second objective, researchers in Mark and Leo’s groups aimed to address a limitation that restricts the potential of Trim-Away to be used to treat disease in patients. In the original Trim-Away strategy, degradation requires the protein targeting molecule to be delivered into the cell by direct injection or by giving cells an ‘electric shock’, which is not practical in patients.
The team demonstrated that a specific part of TRIM21, known as the PRYSPRY domain, is capable of binding small chemical molecules, which may open the way for developing drugs that recruit TRIM21 for therapeutic protein depletion.

“We developed small-molecules that bound to the PRYSPRY domain and chemically linked these to molecules that bind to BRD4, a cancer target, and we have now observed formation of the stable three component (ternary) complexes necessary for therapeutic protein degradation in cells..”
Mark McAlister, Associate Director, Protein Structure & Biophysics, Discovery Sciences, R&D, AstraZeneca
The collaboration continues. A further goal is to test the TRIM21-binding small molecules as inhibitors of TRIM21 mediated neutralisation of viruses with a view to enhancing the efficacy of adenovirus-based vaccines in patients who have adenovirus neutralising antibodies.
Publications and press:
A Method for the Acute and Rapid Degradation of Endogenous Proteins. Clift, D., McEwan, W.A., Labzin, L.I., Konieczny, V., Mogessie, B., James, L.C., Schuh, M. Cell 2017
New technique ‘Trim-Away’ targets and rapidly destroys proteins in cells. Cambridge Network 2017
Quality control in cancer cells
How do cancer cells mark ‘orphaned’ proteins and incomplete protein complexes for degradation?
A Blue Sky funded collaborative project between Manu Hegde, from the LMB’s Cell Biology Division, and Ana Narvaez, then Associate Principal Scientist at AstraZeneca, has uncovered one pathway responsible for disposing of ‘orphan’ proteins during assembly of large multi-protein complexes.
Manu’s group has long been focused on the question of how cells make new proteins, get them to the right place, and fold them into the correct configuration. Protein quality control pathways oversee this process and maintain cell health by degrading proteins that are made incorrectly or are no longer needed. This process is central to various diseases, as failure of quality control is thought to be a key contributor to neurodegeneration. Meanwhile, cancer cells are especially reliant on protein degradation as they produce abnormal amounts of unusable proteins owing to their mutated and rearranged genomes.
This link to disease led to the collaboration with Ana, as her research interest lay in understanding how cancer cells adapt to conditions that might otherwise impede their growth. Manu and Ana decided to investigate how cancer cells boost their quality control pathways, as they posited that this is a crucial adaptation to their increased load of abnormal proteins.

“In cancer, mutations or gene amplifications often cause a change in levels of protein expression and/or stoichiometric imbalance of subunits of protein complexes. This imbalance can be such that some cancers are known to be dependent on the proteosome to survive.”
Ana Narvaez
Ana’s expertise in cancer models helped the team choose cell lines suitable for approaching this problem. Eszter Zavodszky, a senior scientist in Manu’s group, examined protein degradation in breast cancer cells, and subsequently discovered a pathway responsible for recognising failures during the assembly of multi-protein complexes. Because of their aberrant genome, cancer cells are especially likely to produce the subunits of these complexes in incorrect amounts, leading to unassembled ‘orphan’ proteins. Eszter found that when the protein PSMC5, a component of the proteasome, fails to assemble correctly, it is targeted for degradation by the ubiquitin ligase HERC1. Prolonged association of PSMC5 with its assembly chaperone, PAAF1, serves as an indication that its assembly has failed, prompting HERC1 recognition and degradation of orphaned PSMC5.
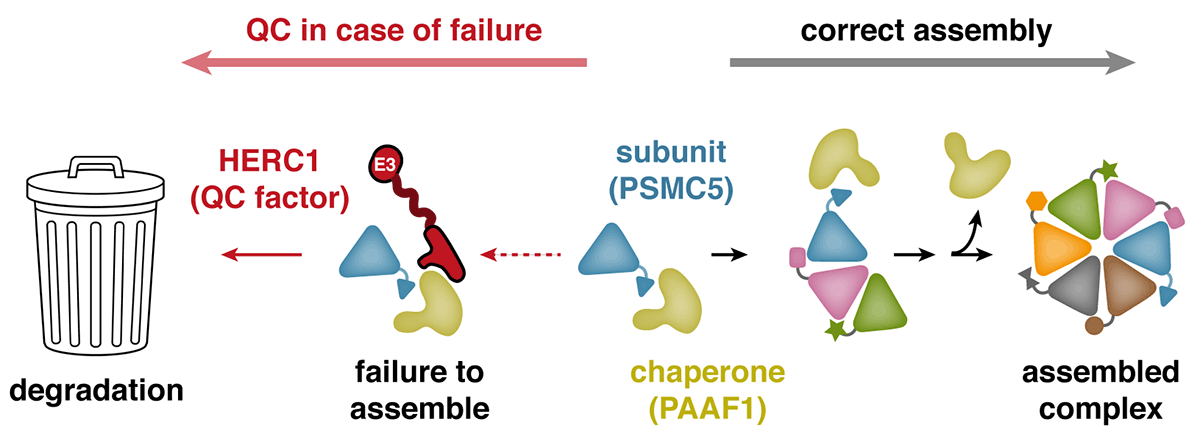
Further analysis revealed that the specific region of HERC1 necessary to recognise PSMC5 is the same region that is mutated in a mouse strain, found decades ago, that develops severe neurodegenerative disease. Eszter found that HERC1 containing this mutation is impaired in recognising the PSMC5-PAAF1 complex. This illustrated the importance of HERC1 in maintaining the correct balance of proteins within cells, particularly in non-dividing cells like neurons, which are especially vulnerable to an accumulation of unassembled or misfolded proteins.

“The central idea that quality control is important to cancer and that its analysis would lead to interesting and new pathways turned out to be valid. And as we had originally suspected, such pathways are not only important in cancer, but also neurodegeneration.”
Manu Hegde, LMB Group Leader
The success of this collaboration has led Manu’s group to redouble their efforts to research protein quality control pathways, and has helped to refine their focus to better understand how cells get rid of ‘orphan’ proteins that are normally part of protein complexes but failed to assemble correctly. ‘We now believe this is one of the major purposes of protein quality control in cells, and working out all the pathways involved for the cell’s most abundant protein complexes will likely be both interesting and important,’ commented Manu.
Publications
Identification of a quality control factor that monitors failures during proteasome assembly. Zavodszky, E., Peak-Chew, S-Y., Juszkiewicz, S., Narvaez, A. J., Hegde, R. S. Science 2021
How do cells respond to surface perturbations?
Identification of proteostatic mechanism triggered by protein aggregation
The Blue Sky collaboration has enabled Harvey McMahon’s group, in the LMB’s Neurobiology Division, to work with Andrew Buchanan, Principal Scientist at AstraZeneca, to study plasma membrane proteostasis. Together, they identified that aggregated cell surface material is internalised by a pathway they named aggregation-dependent endocytosis (ADE).
Previously, research into the mechanisms involved in the response to damage of extracellular proteins has proved difficult to investigate owing to a lack of molecular handles. The BlueSky collaboration helped overcome this, as Andrew Buchanan supplied the biparatopic antibody BS4, which targets the HER2 cell surface receptor in breast cancer cell lines. David Paul, a member of Harvey’s group, was able to use a combination of light microscopy and biochemical assays to study the ADE of BS4.

“It’s been so exciting to partner with excellent scientists, with different expertise and biologic insights. By partnering with Harvey and David we’ve gained a new mechanistic insight into a better trafficking pathway for large molecules. Leveraging this novel insight translationally may pave the way for more effective targeted ADC & gene therapies for a range of diseases. Better press on pushing the boundaries of science to make tomorrows medicines today!”
Andrew Buchanan, Principal Scientist, AstraZeneca
The group found that crosslinking antibodies selectively ‘activate’ the ADE pathway, as do environmental stresses and other molecules which facilitate aggregation of cell surface proteins. After internalisation of the aggregated cell surface proteins by the ADE pathway, the material is then degraded by lysosomes, the primary digestive unit within cells, thereby helping to maintain plasma membrane homeostasis.

“This collaboration with AstraZeneca helped us refine our focus from alternative endocytic routes towards an interest in pathways that may be therapeutically relevant.”
Harvey McMahon, Group Leader, LMB Neurobiology Division

“Key to the success of the project was access to AstraZeneca’s antibody technology. They not only offered continuous intellectual input, but also provided antibodies in amounts that would have been prohibitively expensive if we had to buy them.”
David Paul, Harvey McMahon’s group, LMB Neurobiology Division
Looking forward, Harvey’s group is keen to further investigate ADE to determine if the pathway is implicated in the cell-to-cell spread of neurotoxic amyloid-like protein aggregates, such as tau or alpha-synuclein, which characterise many neurodegenerative diseases. Further understanding of ADE may impact the design of antibody-based therapies, allowing drug developers to maximise efficacy.
Publications
Cell surface protein aggregation triggers endocytosis to maintain plasma membrane proteostasis. Paul, D., Stern, O., Vallis, Y., Dhillon, J., Buchannan, A., McMahon, H. Nature Communications 2023