Scientific Models
The LMB archives hold a small selection of scientific models, the majority of which have been made for or by the LMB. Many of these date back to the early days of the Unit’s protein and DNA research. If you are interested in the LMB’s scientific models and would like to know more, please contact the archivist.
- DNA & Nucleic Acid Models

Skeletal demo model [late 1950’s].
H103cm x W51.5cm x D51.5cm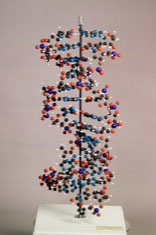
Ball and spoke model [late 1950’s].
‘Portion of macromolecule showing double helical structure’
Cavendish Laboratory [Made by A. Barker].
Scale 2cm = 1Å
H108cm x W46.5cm x D46.5cm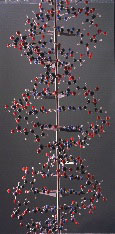
Large ball and spoke model
Built by Alexander Barker, for the International Science Pavilion, Brussels World Exhibition, 1958.
Scale 2.5cm = 1Å
H187cm x W62cm x D62cm, Weight 49.25kg
On display, LMB Library.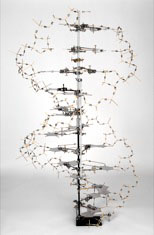
Reproduction of Watson & Crick’s 1953 skeletal model.
Built by Roger Lucke and Claudio Villa, LMB workshop, for the ‘DNA: 50 years of the double helix’ exhibition, Cambridge, 2003.
H200cm x W120cm x D120cm. On display, LMB Reception
Small, modern ball and spoke model
Made by Beevers Models, 2003.
H49cm x W25cm x D25cm
On display, LMB Archive Office
 Base plate from the Watson and Crick 1953 model
Base plate from the Watson and Crick 1953 model
Signed by Francis Crick and Jim Watson.
Donated to LMB by Arthur Arnone (LMB Alumnus)
‘You will notice it is an adenine that is missing its C6-amino nitrogen bond. This is how I found it 31 years ago on a dusty bench [in] the model-building room across from Max’s office. There were about 6 bases there at the time. I had just picked up the adenine when Francis Crick walked by the door on his way down the hall. I rushed over and asked him if this was one of the original bases. He said that he thought it was, and I was bold enough to ask if he would autograph it. A year later I was able to get Jim Watson’s signature when he stopped by the LMB on a visit.’ Arthur Arnone, 28 October 2003.
9 A4 drawings for making atomic models, including proteins and DNA bases, from LMB to Cambridge Repetition Engineers [late 1950’s/early 1960’s].
- Enzyme Models
 ‘Model of the structure of the enzyme, F1-ATPase, which was solved in 1994 by John Walker, joint Nobel prize winner for chemistry 1997, and his colleagues Andrew Leslie, Jan-Pieter Abrahams and Renee Lutter – see Nature370, 621-628 (1994). F1-ATPase is the catalytic component of ATP synthase, the enzyme responsible for the synthesis of ATP in living cells. The gamma subunit (blue) is believed to rotate within the three alpha (red) and three beta (yellow) subunits during catalysis’.The model was constructed by Chris Hellon, Andrew Leslie and Mick Fordham in the MRC LMB workshops.
‘Model of the structure of the enzyme, F1-ATPase, which was solved in 1994 by John Walker, joint Nobel prize winner for chemistry 1997, and his colleagues Andrew Leslie, Jan-Pieter Abrahams and Renee Lutter – see Nature370, 621-628 (1994). F1-ATPase is the catalytic component of ATP synthase, the enzyme responsible for the synthesis of ATP in living cells. The gamma subunit (blue) is believed to rotate within the three alpha (red) and three beta (yellow) subunits during catalysis’.The model was constructed by Chris Hellon, Andrew Leslie and Mick Fordham in the MRC LMB workshops.
On display, outside Max Perutz Lecture Theatre
- Muscle Models

‘A Huxley sliding muscle filament model’ Automated model of muscle filament showing the sliding mechanism.
Hugh Huxley, [Late 1950’s].
- Protein Models and Artefacts
 Early [balsa wood] model of haemoglobin
Early [balsa wood] model of haemoglobin
Max Perutz, 1959.
‘Model of haemoglobin, the protein of the red blood cells. Built by Max Perutz in September 1959 at the Cavendish Laboratory in Cambridge. The segments represent peaks of high density in a three-dimensional map obtained by x-ray crystallography. They outline the course of the four polypeptide chains, while the red disks represent the four haems. This and John Kendrew’s related model of myoglobin were the first protein structures ever solved’.
On display, outside Max Perutz Lecture Theatre
Large wire model of haemoglobin.
Max Perutz [1959]
5.5 Angstrom model of horse haemoglobin
Built in 1959 by Michael Rossmann.
Donated to LMB by Michael Rossmann (LMB Alumnus).
On display, LMB Archive Office
Set of 5.5 Angstrom oxyhaemoglobin maps
Hand-drawn in 1959 by Michael Rossmann.
Donated to LMB by Michael Rossmann (LMB Alumnus).
Sperm Whale Myoglobin
‘Model built by A.A.Barker. To an original design and specification supplied by Dr H.C.Watson, Laboratory of Molecular Biology, Cambridge, England’ [1960’s].
Large ‘sausage’ model of myoglobin.
Built in [1959] by John Kendrew.
On display, LMB Atrium

John Kendrew’s small sausage model of myoglobin, collected from his study at his home in Linton shortly after he died in 1997, with permission of his executor.
Donated to LMB Archives by Michael Fuller, 2005.
Perspex stack of myoglobin showing haem group, in wooden case.
Alpha Helix ball and spoke model
Cavendish Laboratory [1950’s].
Scale 2.5cm = 1Å
On display, LMB Library Mezzanine
Poly(L-glutamine) ball and spoke model of two paired antiparallel β-strands.
Max Perutz [1994]
Reference: Proceedings of the National Academy of Sciences 91: 5355-5358, 1994.
- Ribosome Models and Artefacts

Large glass layered contour map of Ribosome Tetramer: RNA (Opaque) and protein.
W. Kühlbrandt
- Virus Models and Artefacts

Large model of the structure of Tobacco Mosaic Virus (TMV).
Built for the International Science Pavilion, Brussels World Exhibition, 1958, by the sculptor, John Ernest
Rosalind Franklin Group, Birkbeck College, London.
Brought to LMB by Aaron Klug, 1962.
More images

Large model of the structure of Poliomyelitis
Built for the International Science Pavilion, Brussels World Exhibition, 1958, by the sculptor John Ernest.
Rosalind Franklin Group, Birkbeck College, London.
Brought to LMB by Aaron Klug, 1962.
More images

Self-assembling model of the protein shell of a spherical virus particle. Complete model and part model to show the principle of self-assembling.
Further details
Aaron Klug
On display, LMB Archive Office
Two-dimensional paper lattice showing the idea of equivalence in the regular structure of the protein part of virus particles.
Aaron Klug
Collection of geometric models exploring possible virus structure, made from paper, plastics and assorted materials.
Aaron Klug and John Finch
Model to illustrate a portion of the close-packed array formed by helically grooved rods when fully interlocked. It is suggested that in dry tobacco mosaic virus (TMV) the particles pack in this way.” Biochimica et Biophysica Acta19: 403, 1956.
Rosalind Franklin and Aaron Klug
Donated to LMB Archives by Aaron Klug
Small glass layered contour map of Tomato Bushy Stunt Virus [approximately 8 glass layers].
Tony Crowther
Medium glass layered contour map of Tomato Bushy Stunt Virus [approximately 16 glass layers].
Tony Crowther
Medium glass layered contour map of Turnip Crinkle Virus: small particle [approximately 25 glass layers].
Tony Crowther