Scientific Glossary
A glossary of terms used in Insight on Research stories:
- Alternative splicing
- Atomic resolution
- Connectomics
- DNA crosslinks
- History of DNA interstrand crosslinks
- Early Earth conditions
- Endoplasmic reticulum unfolded protein response (UPRER)
- G protein-coupled receptors (GPCRs)
- Innate lymphoid cells, type 2 (ILC2s)
- mTORC1 regulation
- Organoids
- Reverse transcription
- RNA sequencing, single cell
- TATA box binding protein (TBP)
- Transposons
- Tubulins
- Ubiquitination
Alternative splicing
Alternative splicing is a process by which a single gene can be read in slightly different ways to produce multiple versions, or isoforms, of the same protein. Our bodies are able to control this process so that specific versions are made in certain circumstances or in particular cell-types and tissues.
From A new layer of complexity in GPCR signalling could explain differences in receptor function and drug response
 Atomic resolution
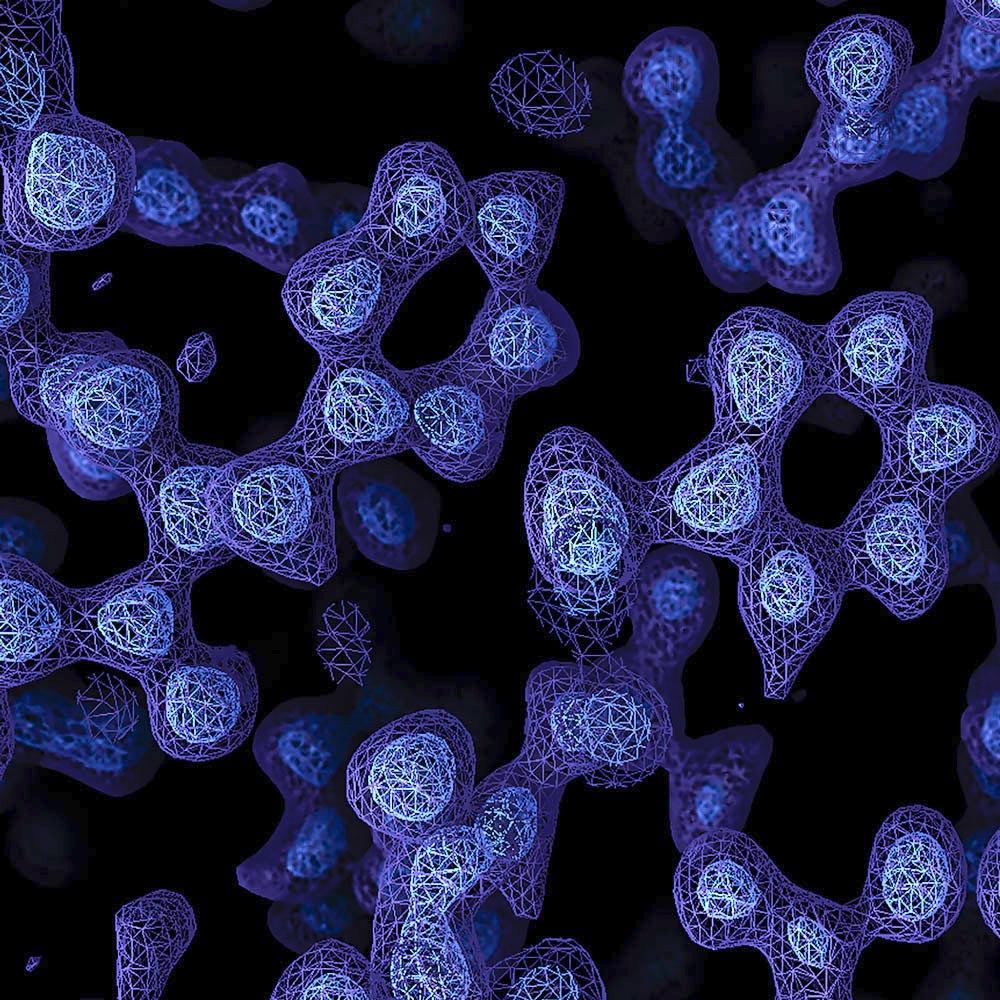
Atomic resolution
Resolution is usually reported in Ångströms, a unit of length that is one ten-billionth of a metre or 0.1 nanometres, ahttps://www2.mrc-lmb.cam.ac.uk/nd refers to the smallest distance between which two objects can be seen to be separate.
The length of a typical carbon-carbon bond is 1.5 Å; other bonds in proteins are a bit shorter. Thus, as the resolution gets down to 1.2 Å, it becomes possible to see individual atoms within a protein, achieving true atomic resolution.
From Atomic advance for cryo-EM
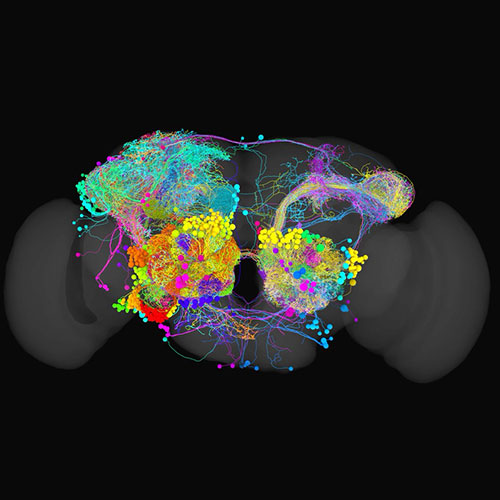
Connectomics
A map shows you what exists in an area and how you can get from one place to another. Connectomics is the study of brain maps identifying every neuron in an organi sm’s nervous system and how they connect with each other.
Mapping a brain is done similarly to manually drawing a road map from satellite images, except in three dimensions using high resolution electron microscopy images of a whole fly brain. This is a time-consuming process, with reconstruction of any individual neuron taking many hours.
From A map to show the way to sense smell, temperature, and humidity
DNA crosslinks
Linkages can form between two nucleotides of DNA within the same strand or between opposite strands of the double-stranded helix, as a result of exposure to exogenous chemicals, such as chemotherapeutic drugs and alcohol, or endogenous by-products of normal cellular metabolism.
These linkages can distort the shape of the DNA helix or prevent two strands from unwinding during DNA replication or transcription. If unrepaired, crosslinks can therefore block DNA from being replicated or transcribed to produce mRNA and then protein. Detection of unrepaired DNA crosslinks can trigger cell death. If crosslinks are found by the FA core complex, DNA repair enzymes are recruited and cell death can be avoided.
From Decade-long collaboration results in the first structure of the Fanconi anaemia core complex
History of DNA interstrand crosslinks
Mustard gas was a deadly chemical weapon first used in World War I against the allies. It killed, maimed and inflicted terror on battlefields near the end of the Great War. By the time of the Second World War, the Allied Forces were anxious about a repeat of the first war. Scientists got busy looking for antidotes to the toxin. This is when two scientists from Yale University, Louis Goodman and Alfred Gilman, discovered that the gas particularly affected white blood cells (which when mutated can develop into cancers), and hypothesised that this agent could be used to treat leukemia. Later on, it was discovered that these and similar anti-cancer drugs worked by generating DNA interstrand crosslinks (ICLs). Since then, agents that induce ICLs are commonly used in chemotherapy. Therefore, understanding ICL repair is important not only to understand the molecular mechanism of Fanconi anaemia, but also DNA repair after chemotherapy.
From How is the Fanconi Anaemia pathway activated to remove DNA lesions?
 Early Earth conditions
Early Earth conditions
How do scientists create early solar system conditions in a lab? UV (mercury) lamps were used for the key photoreduction step, which simulated the early sun’s solar output reaching the surface of Earth, driving key reactions essential for the origin of life. The reactants were dissolved in water, then dried down and heated. This dried mixture is then dissolved in water before being irradiating with UV light, to simulate conditions on a rocky planet like Earth.
From Primitive genetic alphabet based on RNA and DNA
Endoplasmic reticulum unfolded protein response (UPRER)
The endoplasmic reticulum (ER) is a principal site of folding and maturation of many types of proteins. Perturbations to this can lead to accumulation of unfolded proteins, which can lead to disease. The UPRER is a cellular stress response that is activated to deal with this problem. The general aim of the network of signalling pathways that make up the UPRER is to restore or increase the capacity of the ER to fold and process proteins. The capacity of the UPRER to manage this stress decreases in older cells, contributing to age-related diseases, including neurodegenerative diseases.
See A connection between stress detection and lifestyle decisions in worms
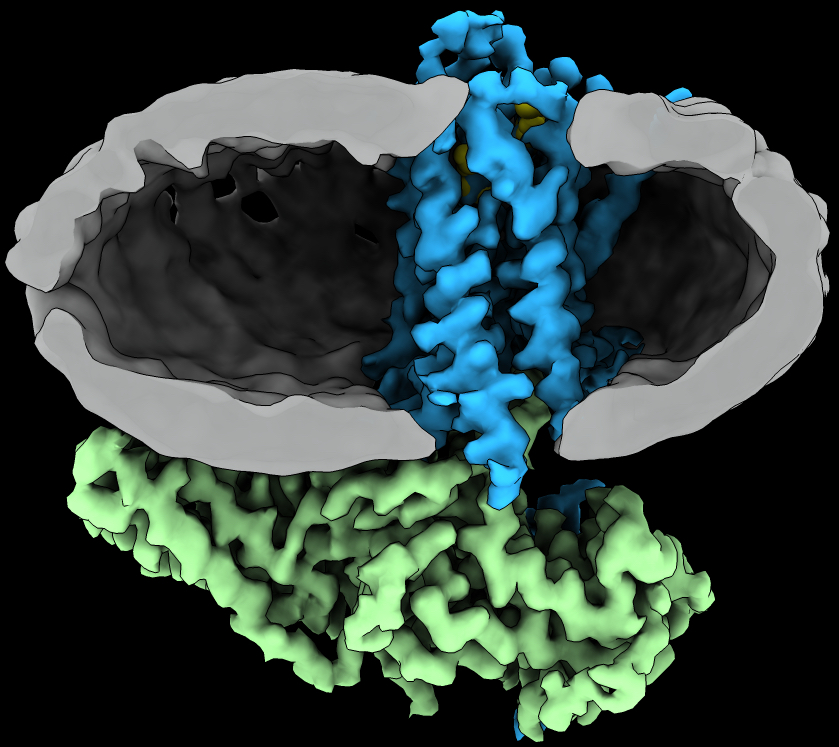 G protein-coupled receptors (GPCRs)
G protein-coupled receptors (GPCRs)
G protein-coupled receptors (GPCRs) are integral membrane proteins that transduce chemical signals from the extracellular matrix into the cell. There are ~800 GPCRs encoded by the human genome and they respond to a diverse array of stimuli such as hormones, neurotransmitters, ions, photons and odorants. They play an important role in physiology and disease, and represent attractive drug targets.
From High-resolution structure of a GPCR-arrestin complex
Innate lymphoid cells, type 2 (ILC2s)
Innate lymphoid cells are thought of as the innate counterparts of T cells. Their vital role in the innate immune response is to secrete effector cytokines that regulate the function of innate and adaptive immune cells. ILC2s localise predominantly at mucosal surfaces where they act as critical sentinels for tissue damage and infection, with a particular role in defence against infection of parasitic worms known as helminths.
From How the fate of immune cell precursors is decided
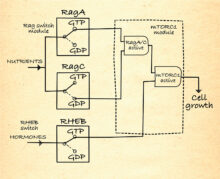
mTORC1 regulation
As depicted in this AND-gate diagram, mTORC1 is only active when the Rags are activated by nutrient abundance, so that mTORC1 is recruited to the lysosomes, and RHEB is activated by hormones. This ensures cell growth only occurs when it is needed and nutrients are plentiful.
From Structural insights into control of cell growth by nutrient availability
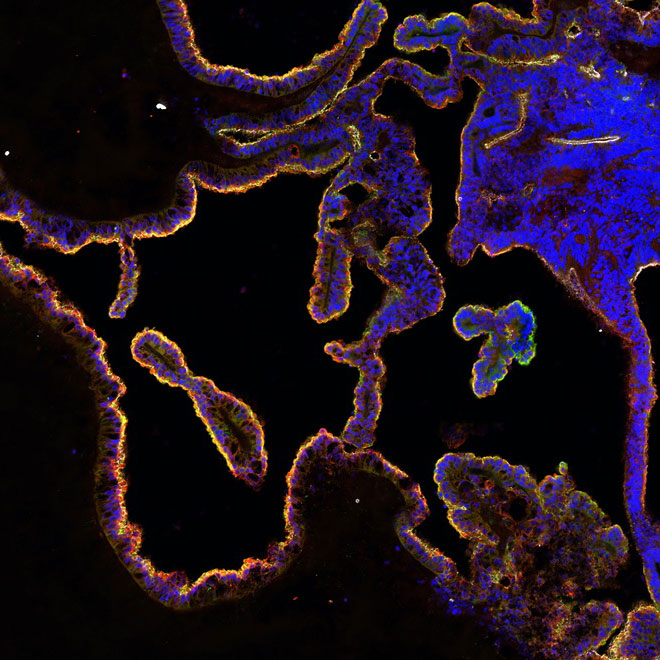 Organoids
Organoids
Organoids are collections of organ-specific cell types that are produced from stem cells and can be studied as miniature, simple versions of organs. Organoids display similar organisation of cell-types to the organ they model and can perform some specific functions.
Scientists produce organoids by directing stem cells towards particular cell-types through the use of signalling molecules, similarly to how the tissues would arise during development.
From Brain organoids with the potential to predict drug permeability
Reverse transcription
The first step of the so-called central dogma of molecular biology is the transcription of DNA to produce RNA. RNA can also be reverse transcribed to produce DNA and synthetic RT enzymes allow access to XNAs.
By enabling scientists to convert RNA to DNA, RT enzymes make it possible for scientists to more easily study what genes are being transcribed inside cells, and therefore which genes are “on”, through core techniques like RT-PCR and RNAseq. Alongside applications in research, this capacity is also used for medical tests, such as to test for the presence of viral RNA, including in COVID-19 tests.
From A new directed evolution technique to unlock the potential of XNAs
RNA sequencing, single cell
By sequencing RNA molecules in cells, scientists can observe which genes are being expressed in those cells. Standard methods of doing this have involved analysis of large populations of cells. However, this may hide rare cell-types and mask small differences between different individual cells.
Single cell RNA sequencing provides gene expression data for individual cells, arming researchers with a deeper understanding of the diversity of cells present in a tissue or organ.
TATA box binding protein (TBP)
TBP recognises and binds to specific DNA sequences (TATA boxes), and bends the DNA. This leads to the recruitment of other factors, such as adapter molecules and the RNA polymerase complex to assemble the pre-initiation complex (PIC) for mediating transcription. TBP is the critical component even in the simplest form of the PIC as seen in single-celled archaea.
From Tracing a transcription initiation factor’s molecular journey across evolutionary timescale
Transposons
Transposons, or transposable elements, are DNA sequences that can change their position within a genome. They act as selfish genetic elements by enhancing their own duplication at the expense of other genes in the same genome, in the same cell.
Retrotransposons are a type of transposon that copy and paste themselves by first transcribing their DNA into RNA and then reverse transcribing that RNA back to DNA. In this way, retrotransposons are very similar to retroviruses like HIV, but, unlike viruses, were not thought to be able to transfer genetic material between cells. Indeed, modern retroviruses and retrotransposons are thought to have a common ancestor.
From Structures of virus-like capsids involved in learning and memory formation
Tubulins
Tubulins are the proteins that join together, or polymerise, to form microtubules. Microtubules control key cellular events such as cell movement, cell shape, and cell division. The dynamics of microtubule polymerisation are highly dependent on the precise levels of tubulin, which probably explains why cells have evolved a mechanism to tightly control its mRNA levels.
Tubulins are critical in the structure and function of neurons and various neurodevelopmental diseases are caused by mutations in different tubulin genes. Tubulin is also the target of drugs used to treat gout and certain types of cancer. The discovery of a factor that regulates tubulin might in the future lead to new therapeutics for these diseases.
From New understanding of protein abundance control
Ubiquitination
Ubiquitin is a small protein that can be attached to other proteins through a process called ubiquitination, which can mark proteins for degradation or regulate activity in a range of other ways. Ubiquitination involves three main steps that are driven by three different groups of enzymes, known as E1s, E2s, and E3s. TRIM21 is an E3 ubiquitin ligase that controls the final step; transfer of ubiquitin from the E2 ubiquitin-conjugating enzyme to the target protein.
From TRIM21 is both enzyme and substrate when creating a signal to degrade bound viruses and proteins