Biophysics

The biophysics facility brings established theories and methods from the physical sciences and applies these to open questions in structural biology. Biophysical techniques contribute to all areas of research in the LMB from simple expression and purification issues through to verifying high-resolution structural information and probing interactions and biology from single molecules through to whole cells.
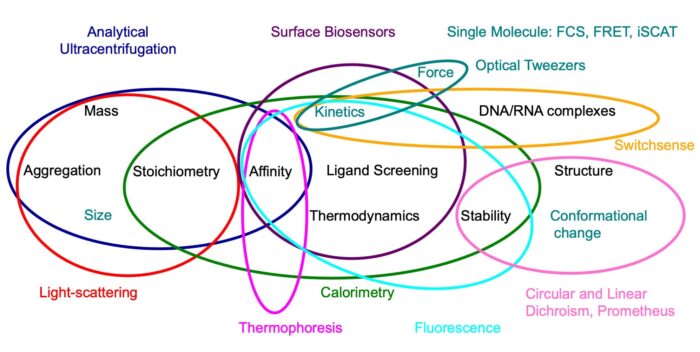
Researchers can access a wide range of biophysical techniques that probe the hydrodynamic, spectroscopic, thermodynamic and physical properties of biological macromolecules and their interactions. Techniques covered include sensor based platforms such as Biacore, Octet and SwitchSense, titration and scanning calorimetry, analytical centrifugation, circular and linear dichroism, fluorescence and scanning fluorimetry, static and dynamic light scattering and microscale thermophoresis. In addition, there are also a number of ‘in-house’ instruments for performing the latest cutting edge techniques in rapid reaction relaxation kinetics and single molecule spectroscopies. Many of these methods are central to the success of research programmes in the LMB.
These world-class facilities have an expert team of scientists headed by Stephen McLaughlin, supported by Chris Batters, who bring many years of experience with these techniques in numerous applications. To help researchers make their own routine measurements, the biophysics team provides an annual lecture series on methods and instrumentation within the LMB as well as offering detailed one-to-one training as required. In addition, these experts can discuss the design of proposed experiments and advise on the interpretation and fitting of results. The team also engages in fully collaborative projects where there is a requirement for specialist skills in the design and execution of the work and where analysis and fitting of data are complex.
In collaboration with AstraZeneca and Imperial College, London, the facility organises an annual one-day symposium on Next Generation Biophysics, that brings together scientists in both academia and industry for stimulating talks examining the application of cutting edge biophysical techniques in complex biological settings.
Selected Papers
Alcón, P., Kaczmarczyk, A.P., Ray, K.K., Liolios, T., Guilbaud, G., Sijacki, T., Shen, Y., McLaughlin, S.H., Sale, J.E., Knipscheer, P., Rueda, D.S., Passmore, L.A. (2024)
FANCD2-FANCI surveys DNA and recognizes double- to single-stranded junctions.
Nature 2024 Jul 31. doi: 10.1038/s41586-024-07770-w.
von Kügelgen, A., Cassidy, C.K., van Dorst, S., Pagani, L.L., Batters, C., Ford, Z., Löwe, J., Alva, V., Stansfeld, P.K., and Bharat, T.A.M (2024)
Membraneless channels sieve cations in ammonia-oxidizing marine archaea.
Nature 630(8015):230-236. doi: 10.1038/s41586-024-07462-5.
Lövestam, S., Li, D., Wagstaff, J.L., Kotecha, A., Kimanius, D., McLaughlin, S.H., Murzin, A.G., Freund, S.M.V., Goedert, M., Scheres, S.H.W. (2024)
Disease-specific tau filaments assemble via polymorphic intermediates.
Nature: 625(7993):119-125. doi: 10.1038/s41586-023-06788-w.
Muir, K.W., Batters, C., Dendooven, T., Yang, J., Zhang, Z., Burt, A., Barford, D. (2023)
Structural mechanism of outer kinetochore Dam1-Ndc80 complex assembly on microtubules.
Science 382(6675):1184-1190. doi: 10.1126/science.adj8736.
Piot, L., Heroven, C., Bossi, S., Zamith, J., Malinauskas, T., Johnson, C., Wennagel, D., Stroebel, D., Charrier, C., Aricescu, A.R., Mony, L., Paoletti, P. (2023)
GluD1 binds GABA and controls inhibitory plasticity.
Science. 382(6677):1389-1394. doi: 10.1126/science.adf3406
Fischer, E.S., Yu, C.W.H., Hevler, J.F., McLaughlin, S.H., Maslen, S.L., Heck, A.J.R., Freund, S.M.V., Barford, D. (2022)
Juxtaposition of Bub1 and Cdc20 on phosphorylated Mad1 during catalytic mitotic checkpoint complex assembly
Nat Commun: 13(1):6381. doi: 10.1038/s41467-022-34058-2.
Nierhaus, T., McLaughlin, S.H., Bürmann, F., Kureisaite-Ciziene, D., Maslen, S.L., Skehel, J.M., Yu, C.W.H., Freund, S.M.V., Funke, L.F.H., Chin, J.W., Löwe, J. (2022)
Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN.
Nat Microbiol: 7(10):1686-1701. doi: 10.1038/s41564-022-01206-9.
Zeng, J., Santos, A.F., Mukadam, A.S., Osswald, M., Jacques, D.A., Dickson, C.F., McLaughlin, S.H., Johnson, C.M., Kiss, L., Luptak, J., Renner, N., Vaysburd, M., McEwan, W.A., Morais-de-Sá E, Clift, D., James, L.C. (2021)
Target-induced clustering activates Trim-Away of pathogens and proteins.
Nat Struct Mol Biol 28(3):278-289.doi: 10.1038/s41594-021-00560-2.
Johnson, C.M. (2021)
Isothermal Titration Calorimetry in Protein-Ligand Interactions: Methods and Applications
Methods in Molecular Biology (2263) 3rd ed. (Daviter, T., Johnson, C.M., McLaughlin, S.H., and Williams, M.A., eds), Humana Press (USA).
Anandapadamanaban, M., Masson, G.R., Perisic, O., Berndt, A., Kaufman, J., Johnson, C.M., Santhanam, B., Rogala, K.B., Sabatini, D.M., Williams, R.L. (2019)
Architecture of human Rag GTPase heterodimers and their complex with mTORC1.
Science: 366(6462):203-210. doi: 10.1126/science.aax3939.
Inglis, A. J., Masson, G. R., Shao, S., Perisic, O., McLaughlin, S. H., Hegde, R. S. and Williams, R. L. (2019)
Activation of GCN2 by the ribosomal P-stalk.
Proc Natl Acad Sci U S A: 116(11):4946-4954. doi: 10.1073/pnas.1813352116. Epub 2019 Feb 25.