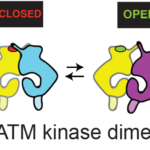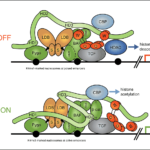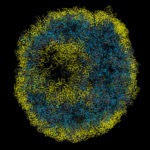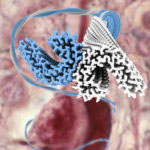
Alzheimer’s disease, the most common neurodegenerative disease, is characterised by the formation of filamentous Tau protein inside nerve cells and amyloid-beta peptides outside cells. Despite more than three decades of research into Tau filaments from a range of different neurodegenerative diseases, atomic structures were still lacking.