
The structures of many proteins have been extensively studied, however it has proved extremely difficult to investigate the structures of the mRNA molecules that carry the genetic information for these proteins.
MRC Laboratory of Molecular Biology
One of the world's leading research institutes, our scientists are working to advance understanding of biological processes at the molecular level - providing the knowledge needed to solve key problems in human health.

Dynactin is a protein complex that activates the dynein motor protein, enabling intracellular transport. It is extremely flexible and has proved very difficult to study by conventional crystallography methods. Now for the first time, research carried out by Andrew Carter and his group in the LMB’s Structural Studies Division, has revealed the structure of this large dynactin complex, using electron cryo-microscopy (cryo-EM).
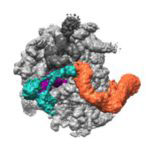
A research team from the LMB’s Cell Biology division, working with colleagues from the Structural Studies division, has revealed how cells are able to find and tag for degradation the partially synthesised proteins generated when ribosomes occasionally stall.
Cells make more than a hundred thousand new proteins every minute. Once in a while, one of the ribosomes making these proteins stalls, leaving an unfinished protein fragment.

When a bacterial cell divides, the cell membrane and cell envelope have to pinch together in the middle of the cell to separate it into two daughter cells. A ring of proteins called the divisome constricts, cleaving the cell in two. The protein FtsZ is a crucial component of this ring and many FtsZ subunits join together in a chain forming long filaments. These FtsZ filaments are anchored to the membrane by another protein, FtsA, so that the membrane also constricts when the FtsZ ring closes.

Recent exciting advances in electron cryo-microscopy (cryo-EM) have allowed scientists to find very detailed structures of some proteins. Still, determining the structure of many proteins remains too difficult for cryo-EM, as the images are too noisy to use for structure determination. Lori Passmore and Chris Russo from the LMB’s Structural Studies Division have designed new specimen support grids, made of pure gold, that improve the microscope image quality.