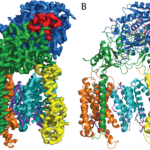
The atomic structure of the 170 kDa membrane protein gamma-secretase, a membrane protein complex that has an important role in Alzheimer’s disease, has been solved using single-particle electron cryo-microscopy (cryo-EM) by Sjors Scheres’ group in the LMB’s Structural Studies Division. This demonstrates for the first time that high-resolution reconstruction of such small molecules can be achieved using cryo-EM.