Since the COVID-19 pandemic began, members of the LMB have found ways to help. From scientifically untangling the virus’s inner workings, providing equipment to hospitals and volunteering for the NHS or as an experienced scientist at a testing centre, our staff and students have joined the response to the COVID-19 pandemic.
This article provides a summary of the contributions of the LMB and its scientists towards the COVID-19 response.
Collaborative scientific efforts

Growing from the collaborative ethos that underpins the LMB’s day-to-day research, our scientific efforts towards the pandemic have been strongly cross-linked, both internally and with other national and international institutes. Our COVID-19 research has been reliant upon collaboration with other leading researchers, including Ravi Gupta at the University of Cambridge, Nick Brindle at the University of Leicester and Bicycle Therapeutics. In line with this, the LMB joined the UK-wide COVID-19 Protein Portal to freely share research and resources in order to facilitate vital research on SARS-CoV-2.
To enable vital research using live SARS-CoV-2 virus by Leo James’s group and others, a team including those with expertise in research safety, building management systems and virus research was rapidly assembled to fast-track opening the LMB’s Containment Level 3 lab.
LMB research on COVID-19 is generally aimed at the following research questions (click on any to find out more about the research being done to address these questions):
How does SARS-CoV-2 infect our cells?
Using the LMB’s expertise in virology and structural and molecular biology, we investigated many aspects of viral function in order to better understand how COVID-19 enters our cells, replicates, and spreads. This could enable developments of new treatments and vaccine strategies.
Groups: John Briggs (moved to MPI August 2021), Andrew Carter, Leo James, Madeline Lancaster, Harvey McMahon, Yorgo Modis, Sean Munro, Julian Sale
See our Insight on Research articles:
SARS-CoV-2 disrupts the brain barrier in human organoids
Furin protease is not essential for SARS-CoV-2 infection
Full list of outputs
Genetic risk factors for death with SARS-CoV-2 from the UK Biobank. (Lu et al.) medRxiv (pre-print – not yet peer-reviewed)
SARS-CoV-2 infects the brain choroid plexus and disrupts the blood-CSF barrier in human brain organoids. (Pellegrini et al.) Cell Stem Cell
Critical care workers have lower seroprevalence of SARS-CoV-2 IgG compared with non-patient facing staff in first wave of COVID19. (Baxendale et al.) Journal of Critical Care Medicine
Furin cleavage of SARS-CoV-2 spike promotes but is not essential for infection and cell-cell fusion. (Papa et al.) PLOS Pathogens
SARS-CoV-2 evolution during treatment of chronic infection. (Kemp et al.) Nature
Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. (Meng et al.) Cell Press
SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. (Mlcochova et al.) Nature
Sequences in the cytoplasmic tail of SARS-CoV-2 spike facilitate expression at the cell surface and syncytia formation. (Cattin-Ortolá et al.) Nature Communications
In vitro evolution predicts emerging CoV-2 mutations with high affinity for ACE2 and cross-species binding. (Bate et al.) bioRxiv (pre-print – not yet peer-reviewed)
Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts tropism and fusogenicity. (Meng et al.) Nature
SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope. (He et al) Nature Microbiology
What is the structure of the SARS-CoV-2 virus?
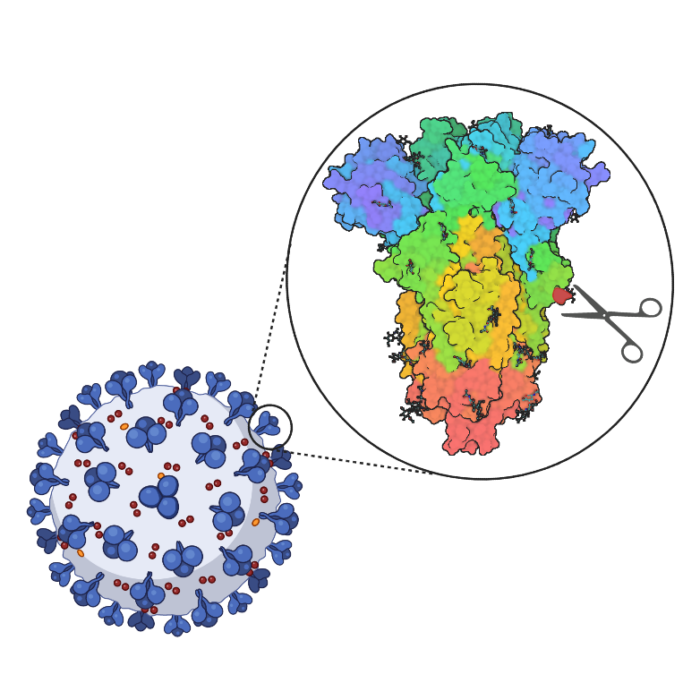
Using electron tomography, LMB researchers have obtained high-resolution structures of parts of the SARS-CoV-2 virus, including the spike proteins, from virus particles. Studying the structure of the virus leads to insights in how the virus functions, both as isolated pathogens and in interactions with antibodies.
Groups: David Barford, John Briggs (moved to MPI August 2021), Andrew Carter, Leo James, Jan Löwe, Chris Russo, Sjors Scheres, John Sutherland
See our Insight on Research articles:
Studying the spike protein of SARS-CoV-2
Targeting RNA replication in SARS-CoV-2
Full list of outputs
A thermostable, closed SARS-CoV-2 spike protein trimer. (Xiong et al.) Nature Structural & Molecular Biology
Structures and distributions of SARS-CoV-2 spike proteins on intact virions. (Ke et al.) Nature
Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. (Naydenova et al.) PNAS
Characterizing flexibility and mobility in the natural mutations of the SARS-CoV-2 spikes. (Panayis et al.) bioRxiv (pre-print – not yet peer-reviewed)
Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike. (Qu et al.) bioRxiv (pre-print – not yet peer-reviewed)
How can we test for, treat and prevent SARS-CoV-2 infection?
Collaborators across the LMB have developed new models, assays, and screening platforms with the goal of investigating new methods and drugs that could be used to test for or treat COVID-19. Research has also focused on why immune responses vary amongst those infected which may help inform the development of further treatments.
Groups: Radu Aricescu, John Briggs (moved to MPI August 2021), Menna Clatworthy, Andres Floto, Julian Gough, Leo James, Jan Löwe, Felix Randow
See our Insight on Research articles:
How immune responses differ between asymptomatic cases and people with severe COVID-19
Testing the capacity for intracellular antibodies to neutralise SARS-CoV-2
Full list of outputs
Combined point-of-care nucleic acid and antibody testing for SARS-CoV-2 following emergence of D614G spike variant. (Mlcochova et al.) Cell Press
Single-cell multi-omics analysis of the immune response in COVID-19. (Stephenson et al.) Nature Medicine
Towards internationally standardised humoral immune correlates of protection from SARS-CoV-2 infection and COVID-19 disease. (Castillo-Olivares et al.) medRxiv (pre-print – not yet peer-reviewed)
Targeting the conserved stem loop 2 motif in the SARS-CoV-2 genome. (Lulla et al.) Journal of Virology
SARS-CoV-2 spike protein stabilized in the closed state induces potent neutralizing responses. (Carnell et al.) Journal of Virology
A functional assay for serum detection of antibodies against SARS-CoV-2 nucleoprotein. (Albecka et al.) EMBO Journal
Single-dose immunisation with a multimerised SARS-CoV-2 receptor binding domain (RBD) induces an enhances and protective response in mice. (Salzer et al.) FEBS Letters
Integrated human/SARS-CoV-2 metabolic models present novel treatment strategies against COVID-19. (Bannerman et al.) Life Science Alliance
Analysis of serological biomarkers of SARS-CoV-2 infection in convalescent samples from severe, moderate and mild COVID-19 cases. (Castillo-Olivares et al.) Frontiers in Immunology
Context-specific emergence and growth of the SARS-CoV-2 Delta variant. (McCrone et al.) Nature
Personal stories and volunteering

We would like to recognise the fantastic contributions made by so many LMB scientists to the COVID-19 response, both in their research and outside of our labs as volunteers helping the NHS, at testing centres, and in their local communities. The following sections detail some of the contributions and include interviews with individuals about their experiences of working through the pandemic.
LMB volunteers
- The LMB assisted with a Cambridge-wide call for volunteers, coordinated by Professor Ian Goodfellow at the Department of Pathology, University of Cambridge.
- 15 LMB-based scientists volunteered at COVID-19 testing centres:
- Tobias Wauer, Elyse Fischer, Hasan Al-Habib, Nadia Cummins, Antonio Galindo, Ewa Gogola, Samuel Lacey, Andrei Mihut, Jack Munns and James Rhodes volunteered as experienced scientists at the COVID-19 National Testing Centre set up by the National Institute of Health Research in Milton Keynes. The 10 volunteers completed 12-hour shifts to help in the national effort to increase COVID-19 testing capacity.
- Tobias Wauer shared his experience of volunteering at the UK Biocentre Lighthouse Lab in The Conversation: ‘My new life as a coronavirus tester’
- Lorena Boquete Vilarino, Shabih Shakeel, Laura Whitworth, Conny Yu and Eszter Zavodszky volunteered as experienced scientists helping to increase local testing capacity for NHS staff at a COVID-19 testing facility located near the LMB on the Cambridge Biomedical Campus.
- The testing volunteers shared details of their experience – see: Putting science skills to new use during the pandemic
- Nina Rzechorzek enrolled as an NHS volunteer and a biomedical researcher in the Cambridge call for volunteers.
- LMB’s Health and Safety department coordinated donations of personal protective equipment (PPE) to Addenbrooke’s Hospital in Cambridge at the start of the pandemic. Following a request from the University of Cambridge, LMB staff assembled a large donation of essential PPE including gloves, disposable aprons and coveralls, safety glasses and overshoes. The collection was coordinated by Jillian Deans, Head of LMB Health and Safety. Jillian said: “It was amazing how quickly LMB staff responded to the request and we hope that the PPE goes some way to help front line NHS staff.”
- Lesley Drynan and Matt Coleman of Biological Services Group coordinated another large effort in getting over 1,500 masks to the University of Cambridge COVID response team that was working with the NHS. The LMB lent its PortaCount mask-fit testing machine to Addenbrooke’s Hospital and Liam Bray trained NHS staff on how to use it.
- Several members of LMB helped their communities in their personal capacities, such as Freda Chapman from our Domestic Services department. Freda signed up to be an NHS volunteer responder and also assisted her neighbour, who is an essential health worker, by providing hot meals.
Personal perspectives of COVID-19 research
We are accompanying our COVID-19 Insight on Research articles with interviews with the scientists who carried out the work, to give some perspective of the experiences of transitioning to COVID-19 research and working at the LMB during the height of the ongoing pandemic.

Xiaoli Xiong
“The institute was dark and quiet probably because I often ended up working until rather late at night. There were very few people working in the institute and I missed finding people to discuss various topics including scientific problems.” Read more…

Zunlong Ke
“As a young researcher, I really hoped that I was able to contribute to the global fight against the pandemic. As it turned out, I was fortunate enough to do so and made important observations of the nature of the virus.” Read more…

Laura Pellegrini
“The challenges, when working with such an infectious pathogen, have been to rethink how to perform certain experiments in order to meet the safety measures in place and this has limited some of the procedures we normally use in the lab.” Read more…

Guido Papa
“I have been studying viruses since my undergraduate degree and, as a virologist, I felt I had to give my contribution to COVID research and join the global fight against the pandemic by putting in place my expertise and knowledge in virology.” Read more…

Kelvin Tuong
“It is exciting to speculate that this work may lead to new therapeutics that could allow individuals to cope better with infection.” Read more…
Images to download
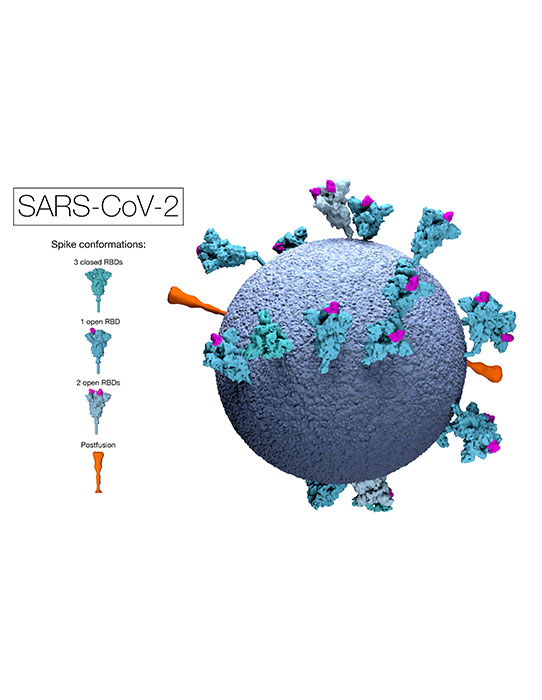
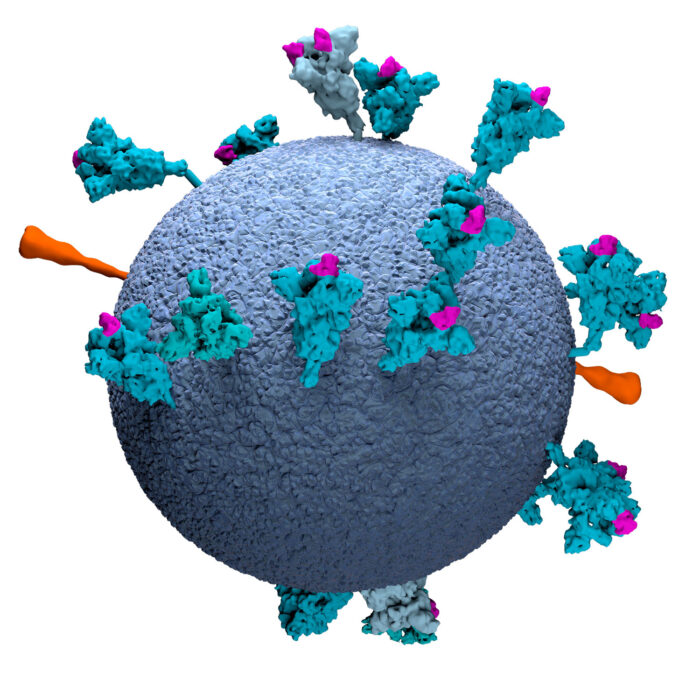
This model of an authentic virus particle reveals positions, conformations and orientations of the spike proteins on the virion membrane (blue). RBD = Receptor Binding Domain.
Image available to download and use under a CC-BY-NC-ND 4.0 International license.
Copyright MRC Laboratory of Molecular Biology.

This graphic clearly shows the positions, conformations and orientations of the spike proteins on the virion membrane (grey).
Image available to download and use under a CC-BY-NC-ND 4.0 International license.
Copyright MRC Laboratory of Molecular Biology.