

Jan Löwe
Prokaryotic cytoskeletons and other molecular machines
jyl@mrc-lmb.cam.ac.ukPersonal group site
Our main area of research is the bacterial cytoskeleton and its roles in bacterial morphogenesis and cell division. In the past, we have been involved in showing that prokaryotes contain both actin and tubulin-like proteins that polymerise into filaments similar to their counterparts in eukaryotes.
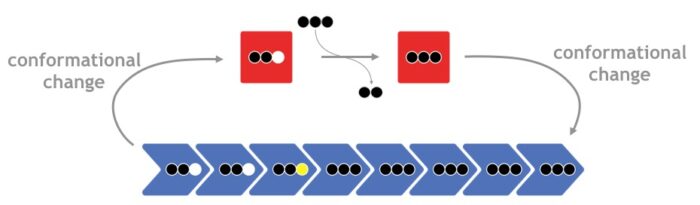
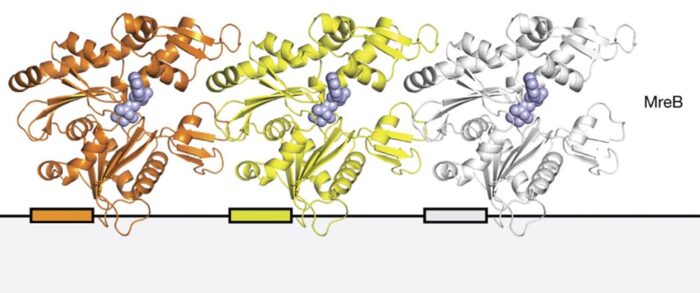
It is now clear that the filament systems can be divided into dynamic, cytomotive and non-dynamic, cytoskeletal. For example, cytomotive filaments of tubulin-like FtsZ are part of the cytokinetic ring around the septum during bacterial cell division. Actin-like cytoskeletal MreB filaments control bacterial cell shape. Bacteria and archaea also contain filament systems that are unrelated to actins and tubulin, and we investigate their roles in morphogenesis and cell division.
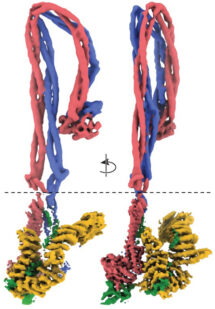
A second theme of the group is SMC complexes, both bacterial and eukaryotic, in the shape of condensins and cohesins. We are currently using cryo-EM in order to discover the precise molecular mechanisms employed by these important and ubiquitous facilitators of chromosome biology.
In order to understand these systems and their mechanisms, in vitro and in vivo, we investigate at the level of cells with fluorescence light microscopy and electron cryo-tomography, and at the level of molecules with X-ray crystallography and electron cryo-microscopy.
Selected Papers
- Deng X., Gonzales Llamazzares A., Wagstaff J., Hale V.L., Cannone G., McLaughlin S.H., Kureisaite-Ciziene D., Löwe J., (2019)
The structure of bactofilin filaments reveals their mode of membrane binding and lack of polarity.
Nature Microbiology 4: 2357-2368. - Bürmann F., Lee B.-G., Hu B., Sinn L., O'Reilly F., Yatskevich S., Rappsilber J., Nasmyth K., Löwe J., (2019)
A folded conformation of MukBEF and cohesin
Nat Struct Mol Biol 26: 227-236. - Wagstaff J., Löwe J., (2018)
Prokaryotic Cytoskeleton - protein filaments organising small cells.
Nature Reviews Microbiology 16: 187-201. - Deng X., Fink G., Bharat T.A.M., He S., Kureisaite-Ciziene D., Löwe J., (2017)
Four-stranded mini microtubules formed by Prosthecobacte BtubAB show dynamic instability.
PNAS 114: E5950-E5958. - Bharat, T.A.M., Murshudov, G.N., Sachse, C. and Löwe, J. (2015)
Structures of actin-like ParM filaments reveal the architecture of plasmid-segregating bipolar spindles.
Nature 523: 106-110. - Gligoris T. G., Scheinost J. C., Bürmann F., Petela N., Chan K-L., Uluocak P., Beckouet F., Nasmyth K. and Löwe J. (2014)
Closing the cohesin ring: structure and function of its Smc-kleisin interface.
Science 346: 963-967.
Group Members
- Danguole Ciziene
- James Collier
- Tina Drobnic
- Liam Gregory
- James Hooker
- Lisa Kashammer
- Martin McAndrew
- Kim Nasmyth
- Leah Pitman
- Lukas Rohland
- Fusinita van den Ent
- James Warren
- Yue Zhang