

Alan Fersht was a Natural Sciences undergraduate and Chemistry postgraduate at Gonville and Caius from 1962-68. He was invited to join LMB as a group leader in 1968 to work on the mechanism of enzymes whose structures were then being solved in the lab and joined a year later after a post-doc at Brandeis with Bill Jencks. He shared room 309 with Max Perutz, who became his mentor. He designed apparatus, which was made in the workshops, to pursue rapid reaction kinetics on enzymes. Alan wrote his book on Enzyme Structure and Mechanism at the end of a bench there with Max’s encouragement between experiments. In 1977, he was appointed Dorothy Hodgkin’s successor as the Wolfson Research Professor of the Royal Society, to be held at Imperial College, where David Blow and Brian Hartley had gone from the LMB. He took up the position after a year’s sabbatical in Arthur Kornberg’s Lab at Stanford to learn gene cloning and DNA replication and proceeded to alienate the organic chemists by the heresy of working on recombinant proteins. In 1981, he began his long collaboration with Greg Winter in developing Protein Engineering, and further alienated the classical enzymologists by working on engineered mutants. In 1988, he was invited back to Cambridge as the Herchel Smith Professor of Organic Chemistry. The MRC, who had supported his work at Imperial College, set him up with the MRC Unit of Protein Function and Design, and almost immediately created the MRC Centre for Protein Engineering (CPE) with Alan as Director and Greg as Deputy Director. The Centre closed and all its staff incorporated into the LMB on his retirement in 2010. Ironically, two of Alan’s students from CPE, Jane Clarke and Sophie Jackson, also became Professors in the Chemistry Department. Both Alan and Greg were knighted for their work on protein engineering and became Masters of Caius and Trinity, respectively, in October 2012, and Jane President of Wolfson in 2018. Alan rejoined the LMB in 2010, where he continued his work on protein folding, misfolding and disease, concentrating on the tumour suppressor p53 until the end of 2017. He was elected FRS in 1983, Honorary Foreign Member of American Academy of Arts and Sciences 5 years later and after another 5 years Foreign Associate National Academy of Sciences (USA), and has been Associate Editor of PNAS since 2000. Among his many prizes have been the Gabor Medal of the Royal Society for Molecular Biology, its Davy Medal for Chemistry and its Royal Medal for his work on protein folding.
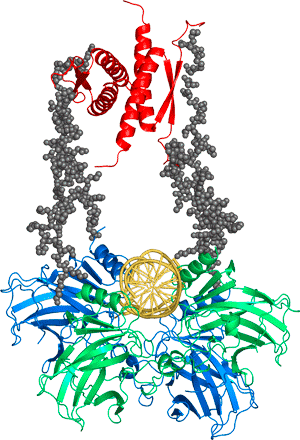
We use an amalgam of protein engineering, structural biology, biophysics and chemistry to study the structure, activity, stability and folding of proteins, and the role of protein misfolding and instability in cancer and disease. We focus on how mutation affects proteins in the cell cycle, particularly the tumour suppressor p53, in order to design novel anti-cancer drugs that function by restoring the activity of mutated proteins.
Cancer is a disease of mutation, and the most commonly mutated protein by far is the tumour suppressor p53. This protein, sometimes called the “Guardian of the Genome”, does not only lead the major defence of the cell against cancer but also has a wide range of roles in the cell cycle, from fertility to senescence. It has a very complicated structure, comprised of two folded and three intrinsically disordered domains in each of its monomers that associate to form dimers and tetramers. It is a major hub protein and interacts with a host of other proteins involved in the cell cycle. We discovered that about 30% of oncogenic mutants of p53 are just temperatures sensitive and can, in theory, be rescued by small molecules.
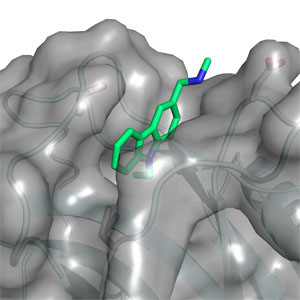
Currently, we have two major projects. “Structural biology of the tumour suppressor p53 and its complexes” in which we are solving the structure of p53 complexed with its partner proteins, such as the Mdm2 and Mdm4, and their complexes. This work is a paradigm for solving the structures of partly disordered proteins, by combining a variety of structural methods.
“Tumour suppressor p53: structure, stability and novel anti-cancer drug development”, funded by an ERC advanced grant, where we are finding the principles of stabilising p53 against denaturation and aggregation by small molecules for the rational design of drugs for rescue of mutants.
Selected Papers
- Wang, G. and Fersht, A.R. (2015)
Mechanism of initiation of aggregation of p53 revealed by Φ-value analysis.
Proc Natl Acad Sci USA. 112: 2437-2442 - Wang, G. and Fersht, A.R. (2015)
Propagation of aggregated p53: Cross-reaction and coaggregation vs. seeding.
Proc Natl Acad Sci USA. 112: 2443-2448 - Joerger, A.C., Bauer, M.R., Wilcken, R., Baud, M.G., Harbrecht, H., Exner, T.E., Boeckler, F.M., Spencer, J. and Fersht, A.R. (2015)
Exploiting Transient Protein States for the Design of Small-Molecule Stabilizers of Mutant p53.
Structure 23: 2246-2255