

Andrew Leslie was a Natural Sciences undergraduate in Cambridge, specializing in crystallography. Following a year in industry, he obtained his PhD from the University of Manchester (UMIST) for structural studies of silicate minerals. His first post-doctoral position was with Struther Arnott at Purdue University in Indiana, using fibre diffraction to determine the structures of a variety of synthetic RNA and DNA polynucleotides. He then changed groups while still at Purdue and worked with Michael Rossmann on southern bean mosaic virus, the first virus structure to be solved in Rossmann’s lab. On returning to the UK, he worked with Alan Wonacott in David Blow’s group at Imperial College on structures of glyceraldehyde 3-phosphate dehydrogenase. A collaboration with Bill Shaw at Leicester University resulted in the structure determination of chloramphenicol acetyl transferase (CAT) that served as a basis for investigating the catalytic mechanism by protein engineering.
In 1988 he came to the LMB, continuing the work on CAT and then, in a collaboration with Robin Carrell in the Haematology department, on ovalbumin as a model for an uncleaved serine protease inhibitor (SERPIN). This was followed by a long-standing collaboration with Sir John Walker (initially in the PNAC division at the LMB but then as head of the MRC Mitochondrial Biology Unit) on structures of F1-ATPase and ATP synthase from both mitochondrial and bacterial sources. During this period, Tony Crowther’s 7Å resolution cryo-EM structure of the hepatitis B viral core particle was used to obtain a crystal structure of the particle at 3.5Å. More recently a collaboration with Chris Tate resulted in the structures of a variety of GPCRs in both inactive and active states, including a complex of a GPCR with a G-protein mimic.
Software development has also been an area of interest, particularly the processing of macromolecular diffraction data, and he continued the development of the MOSFLM program (originally largely the work of Alan Wonacott) for many years. He also had a strong interest in the development of synchrotron macromolecular beam lines and the automation of data collection.
He was elected a fellow of the Royal Society in 2001.
We are involved in projects to determine the atomic structures of macromolecular complexes and membrane proteins using X-ray diffraction.
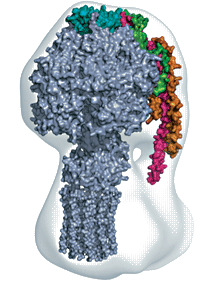
Our work on ATP synthase, in collaboration with Sir John Walker (MRC Mitochondrial Biology Unit) aims to determine the structure of this energy-transducing complex from mitochondria. It is a complex structure of sixteen different polypeptides with a total molecular weight of over 500,000 Da. We have determined the structure of the catalytic sector (F1-ATPase) in a variety of different states. The structures of an F1c10 subcomplex of the yeast ATP synthase, a three-subunit complex of the peripheral stalk, and the NMR structure of another component of the peripheral stalk (the OSCP protein), have been assembled within a cryo-EM derived envelope to provide a model of the whole enzyme. We are now focusing on obtaining high quality crystals of the complete enzyme. We hope that the structure will help us to understand the mechanism that couples proton transport to ATP synthesis.
We are also working on integral membrane proteins, a wide variety of which perform crucial activities in living cells. In collaboration with Chris Tate, we are studying the atomic structures of membrane proteins that are specifically involved in signalling and transport. This includes the G-protein coupled receptors ?1-adrenergic receptor and the adenosine A2A receptor.
We are also developing software (MOSFLM) for processing X-ray diffraction data. Recently this has focussed on a new graphical user interface (GUI) and developments that will allow more automated data collection and processing, especially at synchrotron beamlines. We will be enhancing the algorithms employed by MOSFLM to be able to tackle more challenging problems (such as very weak diffraction, multiple lattices, high mosaicity) as well as optimising processing of images from the new generation of pixel detectors.
Selected Papers
- Powell, H.R., Battye, T.G.G., Kontogiannis, L., Johnson, O. & Leslie, A.G.W. (2017)
Integrating macromolecular X-ray diffraction data with the graphical user interface iMosflm
Nature Protocols 12: 1310-1325. - Carpenter, B., Nehme, R., Leslie, A.G.W. & Tate, C.G. (2016)
Structure of the adenosine A2A receptor bound to an engineered G protein
Nature Communications 536: 104-107. - Leslie, A.G.W., Warne, T. & Tate, C.G. (2015)
Ligand occupancy in crystal structure of β1-adrenergic G protein-coupled receptor
Nature Struct. Mol. Biol. 22: 941-942. - Morales-Rios, E., Montgomery, M.G., Leslie, A.G.W. & Walker, J.E. (2015)
Structure of ATP synthase from Paracoccus denitrificans determined by X-ray crystallography at 4.0 Å resolution
Proc. Natl. Acad. Sci. 112: 13231-13236. - Bason. J.V., Montgomery, M.G., Leslie, A.G.W. and Walker, J.E. (2015)
How Release of Phosphate from Mammalian F1-ATPase Generates a Rotary Sub-step.
Proc. Natl. Acad. Sci. 112: 6009-6014. - Nannenga, B.L., Shi, D., Leslie, A.G.W. and Gonen, T. (2014)
High-resolution structure determination by continuous-rotation data collection in MicroED.
Nature Methods 1: 927-930 - Lebon, G., Warne, T., Edwards, P.C., Bennett, K., Langmead, C.J., Leslie, A.G.W. and Tate, C.G. (2011)
Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation.
Nature 474: 510-514. - Warne, T., Moukhametzianov, R., Baker, J.G., Nehme, R., Edwards, P.C., Leslie, A.G.W., Schertler, G.F.X. and Tate, C.G. (2011)
The structural basis for agonist and partial agonist action on a β1-adrenergic receptor.
Nature 469: 241-244. - Battye, T.G.T., Kontogiannis, L., Johnson, O., Powell, H.R. and Leslie, A.G.W. (2011)
iMosflm: a new graphical interface for diffraction image processing with MOSFLM.
Acta Cryst D67: 271-281.