David Neuhaus studied chemistry at St. Edmund Hall Oxford from 1974-78 and then in 1982 obtained his PhD in synthetic organic chemistry at Imperial College London, working with Prof. Charles Rees. While at Imperial he ran the high-field NMR service for the organic chemistry department; at that time only simple 1D 1H NMR spectroscopy was widely used, so he was able to use most of the out-of-hours spectrometer time to develop his interests in more informative and demanding NMR techniques such as nuclear Overhauser effect experiments. On leaving Imperial he switched full-time to NMR, working with Kurt Wüthrich at ETH Zürich during the period when that group was first developing methodology for solution-phase protein structure determination. His work there included some of the first 1H-detected, heteronuclear experiments on proteins (forerunners of the techniques that today are central to biological NMR structure determination) while studying rabbit metallothionein II, which later turned out to be the first protein structure where NMR substantially corrected an X-ray structure. After another postdoc., this time with Prof. Ray Freeman in Oxford working on NMR method development for applications in organic chemistry, he moved to Cambridge for a short period in the pharmaceutical industry, working at the Parke Davis Research Unit for three years from 1985. Then in 1988 he moved to the LMB to introduce the NMR technique to the lab. During his time at the LMB he has collaborated with many groups both internally and externally on a wide variety of structural problems in diverse research areas, always seeking to identify and exploit those projects that would most benefit from the technique. He is a Fellow of the Royal Society of Chemistry, having chaired their NMR Discussion Group in 2001-2003, has served on a number of international advisory boards in the area of biomacromolecular structures (EBI Macromolecular Structures Group 1999-2002, BioMagResBank 2005-7 (chair in 2007), wwPBD in 2011) and on several editorial boards (Macromolecular Structures, Journal of Magnetic Resonance, Journal of Biomolecular NMR) as well as jointly organising several international conferences and running two invited review journals (Co-ordinating Editor “Quarterly Reviews of Biophysics” 1997-2002, Co-ordinating Editor “Progress in NMR Spectroscopy” 2012-present). Together with Prof. Mike Williamson, he co-authored the standard text on the nuclear Overhauser effect, the phenomenon that underlies the ability of NMR to solve three-dimensional structures (D. Neuhaus and M.P. Williamson, “The Nuclear Overhauser Effect in Structural and Conformational Analysis”, 1st edition VCH 1989, 2nd edition Wiley 2000).
Nuclear magnetic resonance (NMR) spectroscopy is the most powerful physical method available for the study of molecular structure in the solution state, probing interactions between individual atomic nuclei to reveal their spatial relationships and dynamic properties. During my time at the LMB, the technique has become steadily more sensitive and widely applicable, as the field strengths available to us improved from our first 500 MHz spectrometer in 1989 to the 800 MHz models we have today; in conjunction with the introduction of probes using cryogenic receiver coils, this has improved the sensitivity through more than 10-fold over this period. By combining state-of-the-art, multi-dimensional NMR experiments with sample preparation strategies that employ carefully devised labelling patterns of stable isotopes (e.g. 13C, 15N, 2H), we have been able to study detailed three-dimensional solution structures and dynamics in many important biological systems.
A particular strength of NMR is its ability to reveal the structural details of specific interactions between biological macromolecules, especially proteins, and their targets in solution; this is often of crucial importance in understanding the underlying basis of function. We have used NMR to determine structures and interactions in a wide variety of highly challenging systems, starting in the early 1990’s with different types of zinc finger domains from transcription factors (oestrogen receptor and SWI5). Work on DNA and RNA recognition by proteins has been a theme of much of the work since then, including particularly the first NMR structures of protein-RNA complexes, determined in collaboration with Gabriele Varani and Kiyoshi Nagai. Other projects have included work in the areas of protein folding, nuclear trafficking, amoeboid motility, the F1ATPase system, RNA splicing, chromatin targeting, vesicle trafficking, mating type determination in Dictyostelium discoides, and, most recently, intracellular immunity.
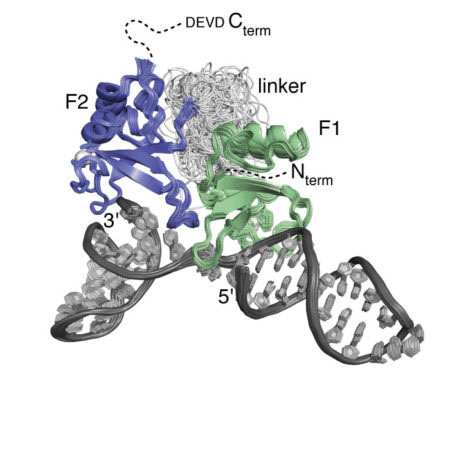
A focus over the past several years has been work designed to unravel the structural basis of the mechanism of the human DNA repair factor PARP-1, which is an important target in cancer therapy. This fascinating enzyme surveys the genome for DNA single-strand breaks, upon detection of which it synthesises and attaches to nearby proteins (including itself) the highly negatively charged, NAD-derived polymer poly(ADP ribose), which in turn acts as a signal for assembly of downstream repair factors. Using a combination of NMR, X-ray crystallography and other biophysical techniques, and in collaboration with groups in Montréal and AstraZeneca, we have been able to disentangle the way in which PARP-1 recognises single-strand breaks, and how this causes a domain assembly cascade within the protein-DNA complex that triggers activation.
Selected Papers
- Ogden, T.E.H., Yang, J.-C., Schimpl, M., Easton, L.E., Underwood, E., Rawlins, P.B., McCauley, M.M., Langelier, M.-F., Pascal, J.M., Embrey, K.J., Neuhaus, D. (2021)
Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acid Research 49: 2266-2288. - Suskiewicz, M. J., Zobel, F., Ogden, T. E. H., Fontana, P., Ariza, A., Yang, J.-C., Zhu, K., Bracken, L., Hawthorne, W. J., Ahel, D., Neuhaus, D., Ahel, I. (2020)
HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation
Nature 579: 598-602. - Eustermann, S.E., Wu, W.F., Langelier, M.F., Yang, J.C., Easton, L.E., Riccio, A.A., Pascal, J.M. and Neuhaus, D. (2015)
Structural basis of detection and signalling of DNA single-strand breaks by human PARP-1
Molecular Cell 60: 742-754 - Eustermann, S., Yang, J.-C., Law, M.J., Amos, R., Chapman, L.M., Jelinska, C., Garrick, D., Clynes, D., Gibbons, R.J., Rhodes, D., Higgs, D.R. and Neuhaus, D. (2011)
Combinatorial Readout of Histone H3 Modifications Specifies Localisation of ATRX to Heterochromatin
Nature Structural and Molecular Biology 18: 777-782. - van Roon, A.M., Loening, N.M., Obayashi, E., Yang, J.-C., Newman, A.J., Hernandez, H., Nagai, K. and Neuhaus, D. (2008)
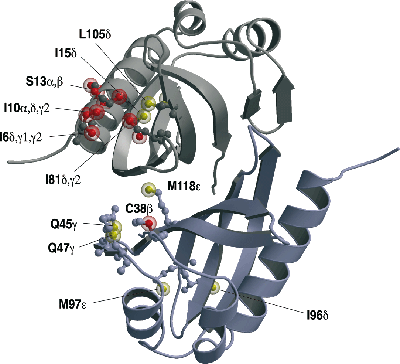
Solution structure of the U2 snRNP protein Rds3p reveals a knotted zinc-finger motif
Proc. Natl. Acad. Sci. U S A 105: 9621-9626. - Kulczyk, A.W., Yang, J.-C. and Neuhaus, D. (2004)
Solution structure and DNA binding of the zinc-finger domain from DNA ligase IIIα
J. Mol. Biol. 341: 723-738. - Allain, F.H.-T., Gubser, C.C., Howe, P.W.A., Nagai, K., Neuhaus, D. and Varani, G. (1996)
Specificity of ribonucleoprotein interaction determined by RNA folding during complex formation
Nature 380: 646-650. - Schwabe, J.W.R., Neuhaus, D. and Rhodes, D. (1990)
Solution structure of the DNA-binding domain of the oestrogen receptor
Nature 348: 458-461.