

Greg Winter studied Natural Sciences at the University of Cambridge. He then completed his PhD at the LMB, working on the amino acid sequence of tryptophanyl tRNA synthetase from the bacterium Bacillus stearothermophilus. His research career has been based almost entirely at the LMB and the MRC Centre for Protein Engineering (CPE). He became a Programme Leader in 1981, was Joint Head of the PNAC Division from 1994-2006, Deputy Director of the LMB from 2006-2011 and acting Director 2007-2008. He was also Deputy Director of CPE from 1990 until its closure in 2010.
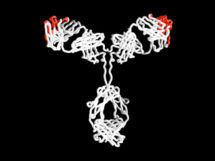 His main research focus is genetic and protein engineering. In his early research Greg was interested in the idea that all antibodies have the same basic structure, with only small changes making them specific for one target. He pioneered techniques in humanised and human therapeutic antibodies, which led to antibody therapies for cancer and diseases such as rheumatoid arthritis and multiple sclerosis. He has established hugely successful spin out companies including: Cambridge Antibody Technology (acquired by AstraZeneca), Domantis (acquired by GlaxoSmithKline) and Bicycle Therapeutics.
His main research focus is genetic and protein engineering. In his early research Greg was interested in the idea that all antibodies have the same basic structure, with only small changes making them specific for one target. He pioneered techniques in humanised and human therapeutic antibodies, which led to antibody therapies for cancer and diseases such as rheumatoid arthritis and multiple sclerosis. He has established hugely successful spin out companies including: Cambridge Antibody Technology (acquired by AstraZeneca), Domantis (acquired by GlaxoSmithKline) and Bicycle Therapeutics.
He is a Fellow of Trinity College, Cambridge and was the Master of Trinity from 2012 to 2019. He was elected a member of the European Molecular Biology Organisation in 1987, a Fellow of the Royal Society in 1990 and Fellow of the Academy of Medical Sciences in 2006, as well as being a Fellow or Honorary Fellow of many other professional organisations. He has been awarded numerous prizes and medals, including the 2018 Nobel Prize for Chemistry. He received a Knighthood for services to Molecular Biology in 2004.
 Natural peptides are flexible molecules; not only does this limit their binding affinity but it facilitates their cleavage by proteases, and it has compromised their development as pharmaceuticals. Our research has aimed to create a new generation of therapeutic agents, “bicycles”, based on highly constrained peptides with a bicyclic structure and a chemical core.
Natural peptides are flexible molecules; not only does this limit their binding affinity but it facilitates their cleavage by proteases, and it has compromised their development as pharmaceuticals. Our research has aimed to create a new generation of therapeutic agents, “bicycles”, based on highly constrained peptides with a bicyclic structure and a chemical core.
We undertook selections to make bicyclic peptides against various targets. In our early research, we described a bicyclic inhibitor of kallikrein. This peptide, PK15, was derived from a large bicyclic random peptide repertoire displayed on filamentous bacteriophage that had been selected against kallikrein. Further studies of PK15 suggested that the structure of the peptide backbone in PK15 must be rather constrained and showed that the amino acid loops of PK15 are constrained in an open “butterfly” configuration in solution.
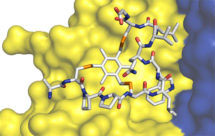
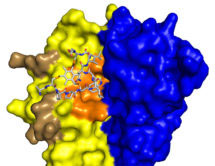 More recently, we made bicyclic peptides against UK18 bicyclic peptide, an inhibitor of human urokinase-type plasminogen activator (uPA) and M21, a ligand for a cryptic epitope of human tumour necrosis factor (TNF). Analysis by X-ray crystallography of the structure of these peptides in complex with their targets revealed that bicyclic peptides have the ability to mimic both antibody and chemical ligands and confirms that bicyclic peptides may be able to combine features of both major classes of pharmaceuticals.
More recently, we made bicyclic peptides against UK18 bicyclic peptide, an inhibitor of human urokinase-type plasminogen activator (uPA) and M21, a ligand for a cryptic epitope of human tumour necrosis factor (TNF). Analysis by X-ray crystallography of the structure of these peptides in complex with their targets revealed that bicyclic peptides have the ability to mimic both antibody and chemical ligands and confirms that bicyclic peptides may be able to combine features of both major classes of pharmaceuticals.
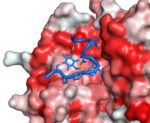 In summary, bicycles can be selected with high affinity and specificity to targets, are highly resistant to attack by proteases and appear to have constrained structures with the two loops in an open “butterfly” configuration. Key patents have been filed, the methodologies for creating bicycles improved and the know-how successfully spun out into the start-up company Bicycle Therapeutics. Bicycles seem able to mimic the properties of both antibodies and small chemical ligands, and may prove very suitable targeting entities for bringing chemical toxins to tumour cells. Such bicycle drug conjugates (BDCs) are currently in development as cancer therapeutics by Bicycle Therapeutics for clinical application.
In summary, bicycles can be selected with high affinity and specificity to targets, are highly resistant to attack by proteases and appear to have constrained structures with the two loops in an open “butterfly” configuration. Key patents have been filed, the methodologies for creating bicycles improved and the know-how successfully spun out into the start-up company Bicycle Therapeutics. Bicycles seem able to mimic the properties of both antibodies and small chemical ligands, and may prove very suitable targeting entities for bringing chemical toxins to tumour cells. Such bicycle drug conjugates (BDCs) are currently in development as cancer therapeutics by Bicycle Therapeutics for clinical application.
Selected Papers
- Luzi, S., Kondo, Y., Bernard, E., Stadler, L. K., Vaysburd, M., Winter, G. and Holliger, P. (2015)
Subunit disassembly and inhibition of TNFα by a semi-synthetic bicyclic peptide.
Protein Eng Des Sel 28: 45-52 - Angelini, A., Cendron, L., Chen, S., Touati, J., Winter, G., Zanotti, G., and Heinis, C. (2012)
Bicyclic peptide inhibitor reveals large contact interface with a protease target.
ACS Chem Biol 7: 817-821 - Baeriswyl, V., Rapley, H., Pollaro, L., Stace, C., Teufel, D., Walker, E., Chen, S., Winter, G., Tite, J. and Heinis, C. (2012)
Bicyclic peptides with optimized ring size inhibit human plasma kallikrein and its orthologues while sparing paralogous proteases.
ChemMedChem 7: 1173-1176 - Heinis, C., Rutherford, T., Freund, S. and Winter, G. (2009)
Phage-encoded combinatorial chemical libraries based on bicyclic peptides.
Nat Chem Biol 5: 502-507