

Murray Stewart initially came to the LMB as a postdoc in Hugh Huxley’s group in 1973, after he had obtained his PhD and worked for two years at the University of New South Wales on the structure of carbon fibres. Murray worked for three years with Hugh, primarily on the structure of tropomyosin and its interactions in muscle thin filaments, before returning to Australia to join the CSIRO Division of Computing Research in Canberra, where he worked primarily on computer image processing and its application to Biological problems and reached the rank of Principal Research Scientist. In 1981 Murray returned to the LMB as a group leader, working initially on muscle thick filament structure and then cell motility before moving to concentrate primarily on the molecular mechanism of nucleocytoplasmic transport. He was elected to membership of the European Molecular Biology Organization in 2006.
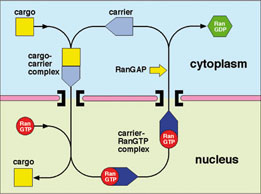
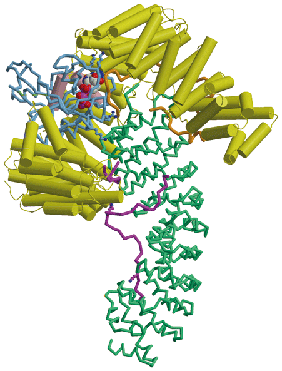 Proteins and nucleic acids are transported between the cytoplasm and nucleus through nuclear pores, huge disc-shaped macromolecular assemblies about 125nm in diameter that perforate the nuclear envelope. Cargo macromolecules are transported through the pores by specific carrier molecules that are able to overcome the barrier function generated by unfolded regions of nuclear pore proteins that fill the transport channel and this process is often orchestrated by the Ran GTPase. Nuclear transport is an example of a Brownian ratchet in which movement through the pores is mediated by Brownian motion, whereas overall transport is effected by transport factors binding cargo selectively in one compartment and releasing it in the other. My research concentrates on understanding how the nuclear transport machinery functions at the molecular level by using a combination of structural, molecular and cellular methods to establish the structure of the proteins involved and how they interact with one another to generate transport. X-ray crystallography, NMR and EM are used to obtain the structure of the components and their interaction interfaces, and then this information is used to generate mutants that can address the functional importance of each interaction. Using this approach, the structures of a range of karyopherin family transport factors together with their complexes with nuclear pore proteins (nucleoporins), cargoes and Ran have been established and used to engineer specific mutants that have identified the importance of karyopherin flexibility that enables RanGTP binding to alter their affinity for cargos.
Proteins and nucleic acids are transported between the cytoplasm and nucleus through nuclear pores, huge disc-shaped macromolecular assemblies about 125nm in diameter that perforate the nuclear envelope. Cargo macromolecules are transported through the pores by specific carrier molecules that are able to overcome the barrier function generated by unfolded regions of nuclear pore proteins that fill the transport channel and this process is often orchestrated by the Ran GTPase. Nuclear transport is an example of a Brownian ratchet in which movement through the pores is mediated by Brownian motion, whereas overall transport is effected by transport factors binding cargo selectively in one compartment and releasing it in the other. My research concentrates on understanding how the nuclear transport machinery functions at the molecular level by using a combination of structural, molecular and cellular methods to establish the structure of the proteins involved and how they interact with one another to generate transport. X-ray crystallography, NMR and EM are used to obtain the structure of the components and their interaction interfaces, and then this information is used to generate mutants that can address the functional importance of each interaction. Using this approach, the structures of a range of karyopherin family transport factors together with their complexes with nuclear pore proteins (nucleoporins), cargoes and Ran have been established and used to engineer specific mutants that have identified the importance of karyopherin flexibility that enables RanGTP binding to alter their affinity for cargos.
My current research concentrates on two main areas. The first is to address the molecular mechanism of the nuclear export of mRNA that does not employ karyopherin-family transport factors or Ran. I am concentrating on understanding the mechanism by which export-competent mRNPs are generated in the nucleus and how they are disassembled in the cytoplasm after passage through the pore. The second area is to investigate the role of nuclear trafficking components, the TREX-2 complex, and the polyA-binding protein, Nab2p, in orchestrating the gene expression machinery, especially in the context of their acting as scaffolds for the formation of the large macromolecular complexes involved in transcription, splicing, mRNA processing and nuclear export.
Selected Papers
- Aibara, S., Gordon, J.M., Riesterer, A.S., McLaughlin, S.H and Stewart M. (2017)
Structural basis for the dimerization of Nab2 generated by RNA binding provides insight into its contribution to both poly(A) tail length determination and transcript compaction in Saccharomyces cerevisiae.
Nucleic Acids Res 45: 1529-1538. doi: 10.1093/nar/gkw1224. - Jani, D., Valkov, E. and Stewart, M. (2014)
Structural basis for binding the TREX2 complex to nuclear pores, GAL1 localisation and mRNA export
Nucleic Acids Res 42: 6686-6697 - Jani, D., Lutz, S., Hurt, E., Laskey, R.A., Stewart, M. and Wickramasinghe, V.O. (2012)
Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export
Nucleic Acids Res 40: 4562-4573 - Ellisdon, A.M., Dimitrova, L., Hurt, E. and Stewart, M. (2012)
Structural basis for the assembly and nucleic acid binding of the TREX-2 transcription-export complex.
Nature Struct Mol Biol 19: 328-336. - Jani, D., Lutz, S., Marshall, N.J., Fischer, T., Köhler, A., Ellisdon, A.M., Hurt, E. and Stewart, M. (2009)
Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export.
Molecular Cell 33: 727-737. - Lee, S.J., Matsuura, Y., Liu, S.M. and Stewart, M. (2005)
Structural basis for nuclear import complex disassembly by RanGTP.
Nature 435: 693-696. - Matsuura, Y. and Stewart, M. (2004)
Structural basis for the assembly of a nuclear export complex.
Nature 432: 872-877.