

As a crystallographer, he has always found it valuable to collaborate with other groups with complementary expertise in biochemistry and cell biology, since the structure alone is not sufficient to understand function. He also continues his long-term interest in software developments in the techniques of X-ray crystallography, mainly in the initial processing of diffraction data, and has been involved in the UK-wide (and international) crystallographic software collaboration CCP4 since its inception in 1979.
He was elected a member of the European Molecular Biology Organisation in 2001 and a Fellow of the Royal Society in 2005.
Macromolecules are moved into cells and between cellular compartments by the movement of membrane vesicles containing protein and lipid cargo. The formation of these vesicles involves the assembly of complex protein machinery to invaginate the membrane, typically forming a polyhedral coat of clathrin around the vesicle.
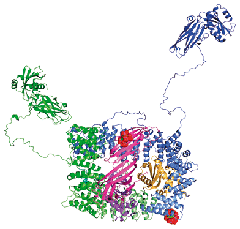
We have been studying the structure and function of protein components of the clathrin-mediated endocytosis and other related systems, using X-ray crystallography, in collaboration with Harvey McMahon and with David Owen (Department of Clinical Biochemistry, Cambridge). This has led to a complete structure of one of the major players, the heterotetrameric AP2 adaptor complex, as well as parts of other components.
As well as protein-protein interactions, the endocytic system involves interactions between soluble proteins and the membrane. A number of endocytic proteins bend membranes into the tight curvature required for the formation of vesicles, and we have been investigating how this is done: for example, BAR domains have an intrinsic curved shape and also insert amphipathic helices into the membrane.
On the basis of the structures, mutants can be designed and used to probe function both in vitro and in vivo. The aim is to build up a complete structural and functional model of vesicle formation and its regulation, and to understand the difference in the trafficking pathways which run between different cell compartments.
Selected Papers
- Kent, H.M., Evans, P.R., Schäfer, I.B., Gray, S.R., Sanderson, C.M., Luzio, J.P., Peden, A.A. and Owen, D.J. (2012)
Structural basis of the intracellular sorting of the SNARE VAMP7 by the AP3 adaptor complex.
Developmental Cell 22: 1-10. - Ingmar B Schäfer, Geoffrey G Hesketh, Nicholas A Bright, Sally R Gray, Paul R Pryor, Philip R Evans, J Paul Luzio, and David J Owen (2012)
The binding of Varp to VAMP7 traps VAMP7 in a closed, fusogenically inactive conformation.
Nature Structural & Molecular Biology 19: 1300-1309. - Evans, P.R. and Murshudov, G.N. (2013)
How good are my data and what is the resolution?
Acta Cryst. D69: 1204–1214.