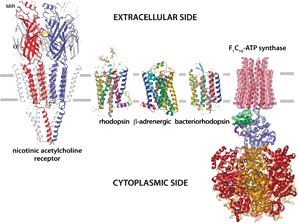
First, many membrane proteins, including most G-protein-coupled receptors, are relatively unstable after purification in detergent: for this class of membrane proteins, Chris Tate and his colleagues in the LMB have recently developed the method of conformational stabilisation (Serrano-Vega et al (2008) PNAS, 105, 877-882) by combining a number of thermostabilising mutants to create much more stable molecules. This has made it possible to tackle structures that were practically impossible previously.
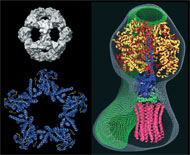
A second problem is that X-ray or electron crystallography needs well-ordered crystals that diffract to high resolution. Since the recent introduction of direct electron detectors and improved computer programs, the resolution of single particle electron cryomicroscopy (cryo-EM) has been greatly improved to the extent that it is now a serious alternative to crystallography. In many but not all cases, atomic structures can now be obtained using single particle cryo-EM. We are therefore working to improve the imaging methods, the computer programs for image processing, and the efficiency of electron detectors, with the goal of realising the full potential of cryo-EM for analysis of single particle structure at atomic resolution.
Selected Papers
- Russo, C.J. & Henderson, R. (2018)
Ewald sphere correction using a single side-band image processing algorithm
Ultramicroscopy 187: 26-33. - Vinothkumar, K.R. & Henderson, R. (2016)
Single particle electron cryomicroscopy: trends, issues and future perspective
Quart. Rev. Biophysics 49: e13, 1-25. - Subramaniam, S., Kuhlbrandt, W. & Henderson, R. (2016)
CryoEM at IUCrJ: a new era
IUCrJ 3: 3-7. - Chen, S., McMullan, G., Faruqi, A.R., Murshudov, G.N., Short, J.M., Scheres, S.H. & Henderson, R. (2013)
High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy.
Ultramicroscopy 135: 24-35. - Henderson, R., Chen, S., Chen, J.Z., Grigorieff, N., Passmore, L.A., Ciccarelli, L., Rubinstein, J.L., Crowther, R.A., Stewart, P.L. and Rosenthal, P.B. (2011)
Tilt-pair analysis of images from a range of different specimens in single particle electron cryomicroscopy.
J Mol Biol 413: 1028-1046. - Vinothkumar, K.R. and Henderson, R. (2010)
Structures of Membrane Proteins.
Quarterly Reviews of Biophysics 43: 65-158.