

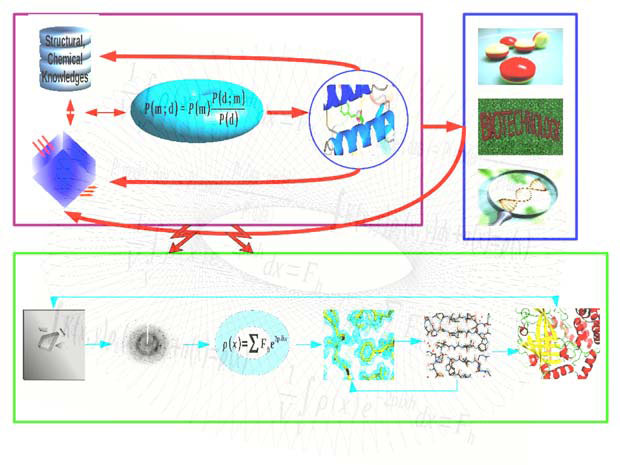
Proteins, nucleic acids and other biological macromolecules take part in virtually all processes within living organisms. Knowledge of their 3-dimensional structures is essential for understanding how they work. Macromolecular crystallography (MX) and Electron cryo-Microscopy (cryo-EM) are powerful experimental techniques that allow the elucidation of 3D structures with high accuracy. According to the Protein Data Bank, around 90% of all structures have been analysed using MX. Due to the resolution revolution in recent years, single particle cryo-EM has become one of the major techniques available for structural biology. Cryo-EM can overcome some of the major limitations of MX, especially for large macromolecular complexes.
Our research is centred on the development of efficient mathematical, statistical, computational algorithms for MX and cryo-EM structure analysis. We implement the developed algorithms in software tools, and distribute them to the structural biology community. Most of our software is distributed to the community via UK-based software initiatives – CCP4 for MX, and CCP-EM for cryo-EM.
We use the Bayesian method, which combines prior structural and chemical knowledge with experimental data, whilst allowing extraction of biologically relevant information from noisy and limited data. Bayesian statistics has two components: the likelihood function, through which information is transferred from the experimental data to the model; and the prior probability distribution, which ensures that the model is consistent with any available knowledge about the system.
Software developed in our group include: a maximum likelihood refinement program – REFMAC5; a molecular graphics program for model building and validation – Coot; a ligand dictionary generator – AceDRG; a structure analysis and restraint generation program – ProSMART; a nucleic acid restraint generation program – LibG; an automatic low-resolution structure refinement pipeline – LORESTR; an automatic structure solution pipeline – BALBES; and a symmetry detection and structure superposition program – ProSHADE.
Selected Papers
- Nicholls, R.A., Tykac, M., Kovalevskiy, O. and Murshudov, G.N. (2018)
Current approaches for the fitting and refinement of atomic models into cryo-EM maps using CCP-EM.
Acta Cryst D74: 492-505. - Kovalevskiy, O., Nicholls, R.A., Long, F., Carlon, A. and Murshudov, G.N. (2018)
Overview of refinement procedures within REFMAC5: utilizing data from different sources.
Acta Cryst D74: 215-227. - Murshudov, G.N. (2016)
Refinement of Atomic Structures Against cyro-EM maps.
Methods in Enzymology, Crowther A ed. 579: 277-305. - Brown, A., Long, F., Nicholls, R.A., Toots, J., Emsley, P. and Murshudov, G. (2015)
Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions.
Acta Cryst D71: 136-153. - Fitzpatrick, A.W., Falcon, B., He, S., Murzin, A.G., Murshudov, G., Garringer, H.J., Crowther, R.A., Ghetti, B., Goedert, M. and Scheres, S.H. (2017)
Cryo-EM structures of tau filaments from Alzheimer’s disease.
Nature 547: 185-190. - Amunts, A., Brown, A., Bai, X.C., Llácer, J.L., Hussain, T., Emsley, P., Long, F., Murshudov, G., Scheres, S.H. and Ramakrishnan, V. (2014)
Structure of the yeast mitochondrial large ribosomal subunit.
Science 343: 1485-1489.
Group Members
- Lucrezia Catapano
- Paul Emsley
- Fei Long
- Martin Maly
- Alexander Shtyrov