

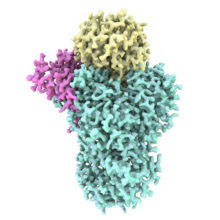
We are studying protein complexes that regulate gene expression. Eukaryotic genes are normally transcribed as pre-mRNAs that must be processed before they are exported from the nucleus and translated into proteins. At the 3′-end, the pre-mRNA is cleaved and a poly(A) tail is added. Several protein factors are required to coordinate the 3´-end processing reactions, including a large multiprotein complex called Cleavage and Polyadenylation Factor (CPF/CPSF).
The poly(A) tail is required for export of the mRNA into the cytoplasm, to enhance mRNA stability and to stimulate translation. Its length can be regulated to influence these functions. For example, shortening of the poly(A) tail can decrease the efficiency of translation to control gene expression, and is the first step in mRNA turnover. Poly(A) tails are shortened by the evolutionarily conserved Ccr4-Not and Pan2–Pan3 complexes. We aim to understand how the CPF, Ccr4-Not and Pan2-Pan3 complexes control poly(A) tail addition and removal.
DNA must be passed on faithfully from cell to cell and across generations to maintain gene expression. DNA crosslinks can occur after exposure to chemicals including chemotherapeutic drugs or alcohol, but also as a result of normal cellular metabolism. These crosslinks block DNA replication and transcription. If crosslinks cannot be repaired, it leads to human disease including Fanconi anemia and cancer. We aim to understand how DNA crosslinks are repaired by the Fanconi anaemia DNA repair pathway.
We are using a hybrid approach to understand the molecular mechanisms of these and other multi-protein complexes. In particular, we use electron cryo-microscopy (cryo-EM), alongside X-ray crystallographic, biophysical, biochemical and genetic techniques. Cryo-EM is rapidly evolving and we are developing new methods to help determine protein structures. Overall, our aim is to establish fundamental principles underlying the assembly of multi-protein complexes, define their structures, gain insight into their activities and regulation, and identify roles for proteins of unknown function.
Selected Papers
- Alcón, P., Kaczmarczyk, A.P., Ray, K.K., Liolios, T., Guilbaud, G., Sijacki, T., Shen, Y., McLaughlin, S.H., Sale, J.E., Knipscheer, P., Rueda, D.S., Passmore, L.A. (2024)
FANCD2-FANCI surveys DNA and recognizes double- to single-stranded junctions
Nature - Rodríguez-Molina, J.B., O'Reilly, F.J., Fagarasan, H., Sheekey, E., Maslen, S., Skehel, J.M., Rappsilber, J., Passmore, L.A. (2022)
Mpe1 senses the binding of pre-mRNA and controls 3' end processing by CPF
Mol Cell. 82(13): 2490-2504 - Shakeel, S.†, Rajendra, E.†, Alcón, P., O’Reilly, F., Chorev, D.S., Maslen, S., Degliesposti, G., Russo, C.J., He, S., Hill, C.H., Skehel, J.M., Scheres, S.H.W., Patel, K.J., Rappsilber, J., Robinson, C.V. and Passmore, L.A. (2019)
Structure of the Fanconi anemia monoubiquitin ligase complex
Nature 575: 234-237 - Tang, T.T.L., Stowell, J.A.W., Hill, C.H. and Passmore, L.A. (2019)
The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases
Nat. Struct. Mol. Biol. 26: 433-442 - Webster, M.W., Chen, Y.H., Stowell, J.A.W., Alhusaini, N., Sweet, T., Graveley, B.R., Coller, J.* and Passmore, L.A.* (2018)
mRNA deadenylation is coupled to translation rates by the differential activities of Ccr4-Not nucleases
Mol. Cell 70: 1089-1100.e8 - Casañal A, Kumar A, Hill CH, Easter AD, Emsley P, Degliesposti G, Gordiyenko Y, Santhanam B, Wolf J, Wiederhold K, Dornan GL, Skehel M, Robinson CV and Passmore LA (2017)
Architecture of eukaryotic mRNA 3′-end processing machinery
Science 358: 1056-1059 - Russo, C.J. and Passmore, L.A. (2014)
Ultrastable gold substrates for electron cryomicroscopy
Science 346: (6215):1377-1380.
Group Members
- Brian Carrick
- Caroline Dean
- Josh Eaton
- Holly Fagarasan
- Marc Fiedler
- Megan Gough
- Xueyan Li
- Alexandra Marti
- Teodora Milanovic
- James Stowell
- Philipp Zuber
- Linyu Zuo