

Information transfer in the nervous system occurs at synapses, where presynaptic signals are interpreted by postsynaptic receptors. We study this process with a focus on AMPA-type glutamate receptors.
AMPA-Rs are glutamate-gated cation channel tetramers. They are the prime mediators of excitatory neurotransmission and are regulators of synaptic plasticity, which underlies higher-order cognitive processes. Our ultimate aim is to understand how AMPA-R signalling contributes to learning at synapses at various levels of complexity.
First, we study fundamental mechanisms underlying AMPA-R signalling with a specific focus on receptor biogenesis and allosteric modulation by interacting proteins. This aspect includes structural approaches such as X-ray crystallography, electron cryo-microscopy (cryo-EM), complemented by simulations of receptor dynamics and high-resolution electrophysiological recordings.
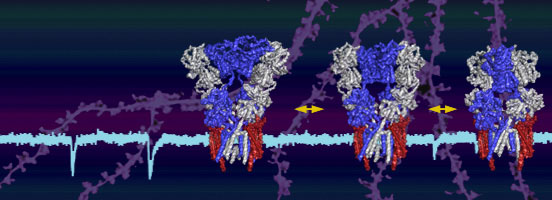
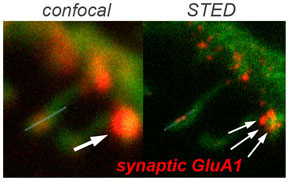
Secondly, we build on this information to unravel AMPA-R operation at synapses. We ask how AMPA-Rs of different subunit compositions are selectively targeted to and anchored at potentiated synapses, and how structural dynamics of AMPA-R tetramers contribute to functional and structural synaptic plasticity. Towards this aim we utilize a combination of brain slice electrophysiology and super-resolution light microscopy approaches.
Selected Papers
- Zhang, D., Ivica, J., Krieger, J.M., Ho, H., Yamashita, K., Stockwell, I., Baradaran, R., Cais, O., Greger, I.H. (2023)
Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor
Nature 621(7980): 877-882 - Zhang, D., Watson, J.F., Matthews, P.M., Cais, O. and Greger, I.H. (2021)
Gating and modulation of a hetero-octameric AMPA glutamate receptor
Nature 594(7863): 454-458. https://doi.org/10.1038/s41586-021-03613-0 - Herguedas, B., Watson, J.F., Ho, H., Cais, O., García-Nafría, J., Greger, I.H. (2019)
Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP γ8
Science 364(6438): pii: eaav9011. doi: 10.1126/science.aav9011. - Watson JF, Ho H, Greger IH. (2017)
Synaptic transmission and plasticity require AMPA receptor anchoring via its N-terminal domain
Elife 14;6: pii: e23024. - Greger, I.H., Watson, J.F., Cull-Candy, S.G. (2017)
Structural and Functional Architecture of AMPA-Type Glutamate Receptors and Their Auxiliary Proteins
Neuron 94: (4):713-730. - Herguedas, B., García-Nafría, J., Cais, O., Fernández-Leiro, R., Krieger, J., Ho, H. and Greger, I.H. (2016)
Structure and organization of heteromeric AMPA-type glutamate receptors
Science 352: (6285):aad3873. - Cais, O., Herguedas, B., Krol, K., Cull-Candy, S.G., Farrant, M. and Greger, I.H. (2014)
Mapping the interaction sites between AMPA receptors and TARPs reveals a role for the receptor N-terminal domain in channel gating.
Cell Rep. 9: 728-40 - Penn, A.C., Balik, A., Wozny, C., Cais, O. and Greger I.H. (2012)
Activity-mediated AMPA receptor remodeling, driven by alternative splicing in the ligand-binding domain.
Neuron 76: 530-10
Group Members
- Ondrej Cais
- Josip Ivica
- Terunaga Nakagawa
- Mikel Perez Rodriguez
- Aditya Pokharna
- Nayanika Sengupta
- Imogen Stockwell
- Fei Wu