

Roger Williams
Structural studies of phosphoinositide signalling and intracellular sorting
rlw@mrc-lmb.cam.ac.ukPersonal group site
Eukaryotic cells have evolved a number of mechanisms to respond to environmental stress. These stress responses are tempered by growth factors and other signals from surrounding cells in multicellular organisms. Our research programme involves the mechanisms whereby these cellular signals are regulate mammalian cells. Phosphoinositide 3-kinases (PI3Ks) modify lipid membranes and the modified phospholipids produced by the enzymes serve as second messengers and sorting signals, and they are important for cell response to environmental cues. PI3Ks are part of a larger family of kinases that also include the PI3K-related protein kinases (PIKKs). We study the structures, dynamics and functions of PIKKs and related pathways in cellular signalling, sorting and nutrition.
Our work has included almost all of the members of the PI3K family: class I PI3Ks activated by receptor tyrosine kinase and GPCRS, the primordial class III PI3K complex present in all eukaryotes, the golgi-associated PI4K and the PIKK known as target of rapamycin (TOR). Each of these enzymes is a regulatory nexus, and our work produces variants in which specific regulatory inputs of the large enzyme complexes are ablated without affecting other activities. These modified complexes provide tools to decipher cellular regulation in ways that would not be possible with other approaches. We use X-ray crystallography, electron cryo-microscopy (cryo-EM) and hydrogen-deuterium exchange mass spectrometry (HDX-MS) to marry structures with dynamics. In collaboration with our pharmaceutical partners, we are seeking to develop HDX-MS approaches to give better insight into conformational dynamics that could result in new therapeutic strategies. On-going efforts in the group concern the structural and dynamic basis of regulation of oncogenic PI3Ks, autophagic PI3K complexes, integration of amino acid and growth factor sensing in the PI3K pathway, regulation of ribosomes in response to amino acid starvation and DNA damage repair.
Selected Papers
- Heimhalt M, Berndt A, Wagstaff J, Anandapadamanaban M, Perisic O, Maslen S, McLaughlin S, Yu CW, Masson GR, Boland A, Ni X, Yamashita K, Murshudov GN, Skehel M, Freund SM, Williams RL (2021)
Bipartite binding and partial inhibition links DEPTOR and mTOR in a mutually antagonistic embrace.
Elife 10: :e68799. PMID: 34519269. - Tremel S, Ohashi Y, Morado DR, Bertram J, Perisic O, Brandt LTL, von Wrisberg MK, Chen ZA, Maslen SL, Kovtun O, Skehel M, Rappsilber J, Lang K, Munro S, Briggs JAG, Williams RL (2021)
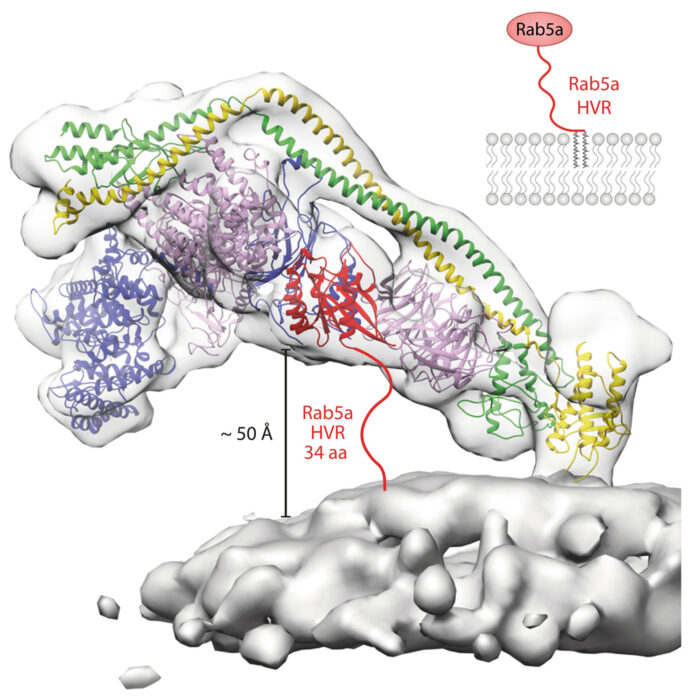
Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes.
Nat Commun.1564. - Anandapadamanaban M, Masson GR, Perisic O, Berndt A, Kaufman J, Johnson CM, Santhanam B, Rogala KB, Sabatini DM, Williams RL. (2019)
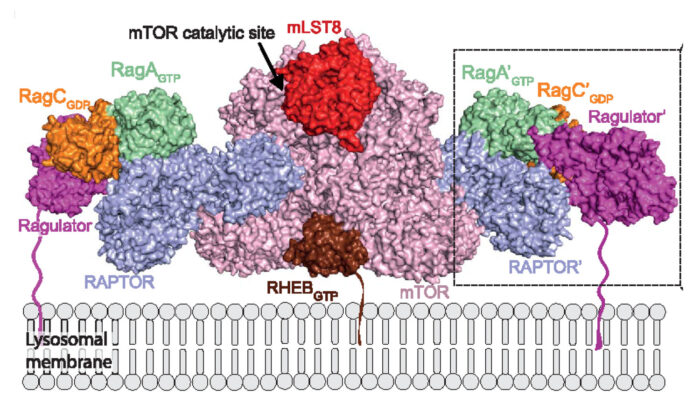
Architecture of human Rag GTPase heterodimers and their complex with mTORC1.
Science 366(6462): 203-210. - Baretić D, Pollard HK, Fisher DI, Johnson CM, Santhanam B, Truman CM, Kouba T, Fersht AR, Phillips C, Williams RL (2017)
Structures of closed and open conformations of dimeric human ATM.
Sci Adv. 10;3(5): e1700933. - Baretić D, Berndt A, Ohashi Y, Johnson CM, Williams RL. (2016)
Tor forms a dimer through an N-terminal helical solenoid with a complex topology.
Nat Commun. 7: 11016. - Rostislavleva, K., Soler, N., Ohashi, Y., Zhang, L., Pardon, E., Burke, JE, Masson, GR, Johnson, C., Jan Steyaert, J., Ktistakis, NT, Williams, RL. (2015)
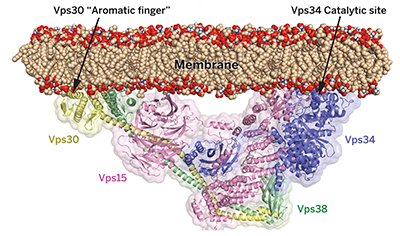
Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes.
Science 350: aac7365. - Burke, JE, Inglis, AJ, Perisic, O,Masson, GR, McLaughlin, SH, Rutaganira, F, Shokat, KM and Williams RL. (2014)
Structures of PI4KIIIβ complexes show simultaneous recruitment of Rab11 and its effectors.
Science 344: 1035-1038. - Angulo I, Vadas O, Garçon F, Banham-Hall E, Plagnol V, Leahy TR, Baxendale H, Coulter T, Curtis J, Wu C, Blake-Palmer K, Perisic O, Smyth D, Maes M, Fiddler C, Juss J, Cilliers D, Markelj G, Chandra A, Farmer G, Kielkowska A, Clark J, Kracker S, Debré M, Picard C, Pellier I, Jabado N, Morris JA, Barcenas-Morales G, Fischer A, Stephens L, Hawkins P, Barrett JC, Abinun M, Clatworthy M, Durandy A, Doffinger R, Chilvers ER, Cant AJ, Kumararatne D, Okkenhaug K, Williams RL, Condliffe A, Nejentsev S. (2013)
Phosphoinositide 3-kinase δ gene mutation predisposes to respiratory infection and airway damage.
Science 342: (6160):866-871.
Group Members
- Madhanagopal Anandapadamanaban
- Maxime Bourguet
- Antoine Dessus
- Grace Gong
- Iain Hay
- Saiful Islam
- Yohei Ohashi
- Olga Perisic