

Our bodies are built from trillions of cells of at least several hundred varieties. To maintain our health, each cell relies on thousands of internal biochemical reactions carried out by around ten thousand different types of proteins. The cell’s proteins (called the proteome) must be made in the correct amount, folded into a precise shape, and placed in the correct location. The process of maintaining a cell’s proteome within normal limits is called proteostasis. How does a cell modulate its proteostatic processes? How are these processes regulated in response to environmental stress? And how are they affected in pathological conditions? Our lab aims to investigate the molecular basis of proteostasis regulation in normal cells and tissues. We further aim to understand how these processes are affected in diseases associated with altered proteostasis, such as cancer or neurodegenerative diseases.
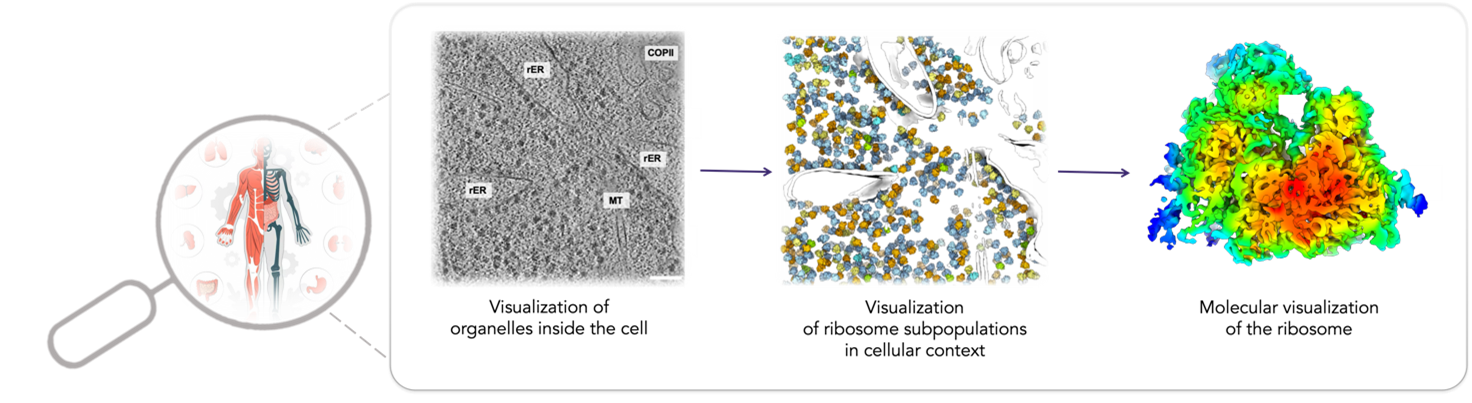
A complex network of molecular machines is tasked with producing the correct amount of properly folded and assembled proteins and making them available at the right location and time. The machines that comprise this proteostasis network include the ribosome, which synthesizes new proteins, chaperones that help new proteins fold, and degradation factors that eliminate incorrect or damaged proteins. To visualize these processes in cells, our lab employs an advanced combination of light microscopy, cryo-electron microscopy, and computational analysis. This interdisciplinary approach will enable a molecular understanding of proteostasis networks in the context of normal and diseased tissues. We expect that our results will reveal insights into pathogenesis and could facilitate new therapeutic approaches for restoring cellular function.
Selected Papers
- Fedry J, Silva J, Vanevic M, Fronik S, Mechulam Y, Schmitt E, des Georges A, Faller W, Förster F (2023)
Visualization of translation reorganization upon persistent collision stress in mammalian cells.
Biorxiv - Gemmer M, Chaillet ML, van Loenhout J, Cuevas Arena R, Vismpas D, Gröllers-Mulderij M, Koh FA, Albanese P, Scheltema R, Howes SC, Kotecha A, Fedry J, Förster F (2023)
Snapshots of translation and protein translocation at the ER membrane.
Nature 614 (7946): 160-167 - Kucinska MK, Fedry J, Galli C, Morone D, Raimondi A, Solda T, Förster F, Molinari M (2023)
SEC62 and TMX4 control asymmetric autophagy of the nuclear envelope upon LINC complex disassembly.
Nature Communications 14 (1): 3497
Group Members
- Prince Chivaka
- Oscar Despard
- Irene Diaz Lopez
- Lovisa Jonsson