

Michael Hastings
Neurons and biological timing: molecular neurobiology of the circadian clock
Personal group site
Circadian rhythms are those daily cycles of physiology and behaviour that persist when organisms are isolated from the external world. They are expressed at all levels of life, from prokaryotic blue-green algae to higher plants and animals. Their biological role is to anticipate and thereby allow organisms to adapt to the solar day and night. In humans the cycle of sleep and wakefulness is the most obvious circadian rhythm, reflecting a profound alternation of brain states. Disruption of our circadian programme through shift work, old age and neurological disease is a significant and growing cause of chronic illness.
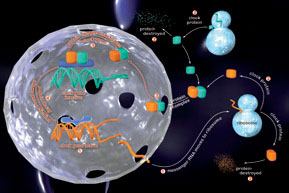
The principal circadian pacemaker is located in the suprachiasmatic nucleus (SCN) of the hypothalamus, where individual neurons can operate as selfsustained circadian clocks. This same clock mechanism is also present in our major organ systems; heart, lungs, liver, kidney etc. The SCN maintains synchrony amongst these sub-ordinate clocks via its control over behaviour, neuroendocrine pathways and the autonomic nervous system. The genes responsible for encoding our circadian clockwork have recently been identified.
We are using real-time in vivo fluorescence and bioluminescence imaging, DNA microarrays, proteomic expression analyses and molecular genetic manipulations to understand how these “clock” genes and their protein products are able to assemble themselves into a 24h time-keeper, and how the central and peripheral timers together co-ordinate our metabolic and physiological rhythms. Through this approach we aim to provide a molecular genetic explanation for one of the most conserved and ancient forms of behaviour-circadian timing.
Selected Papers
- McManus, D., Polidarova, L., Smyllie, NJ., Patton, AP., Chesham, JE., Maywood, ES., Chin, JW., Hastings, MH. (2022)
Cryptochrome 1 as a state variable of the circadian clockwork of the suprachiasmatic nucleus: Evidence from translational switching.
PNAS 119: No. 34. - Smyllie, NJ., Bagnall, J., Koch, AA., Niranjan, D., Polidarova, L., Chesham, JE., Chin, JW., Partch, CL., Loudon, ASI., Hastings, MH. (2022)
Cryptochrome proteins regulate the circadian intracellular behavior and localization of PER2 in mouse suprachiasmatic nucleus neurons.
PNAS 119: No. 4 - Patton, A. P., Edwards, M. D., Smyllie, N. J., Hamnett, R., Chesham, J. E., Brancaccio, M., Maywood, E. S. and Hastings, M. H. (2020)
The VIP-VPAC2 neuropeptidergic axis is a cellular pacemaking hub of the suprachiasmatic nucleus circadian circuit.
Nat Commun 11: 3394. - Brancaccio, M., Edwards, M. D., Patton, A. P., Smyllie, N. J., Chesham, J. E., Maywood, E. S. and Hastings, M. H. (2019)
Cell-autonomous clock of astrocytes drives circadian behavior in mammals.
Science 363: 187-192. - Hastings, MH., Maywood, ES., Brancaccio, M. (2018)
Generation of circadian rhythms in the suprachiasmatic nucleus.
Nat. Rev. Neurosci.19(8):453-469. - Brancaccio M., Patton AP., Chesham JE., Maywood ES., Hastings MH. (2017)
Astrocytes Control Circadian Timekeeping in the Suprachiasmatic Nucleus via Glutamatergic Signaling.
Neuron 93(6): 1420-1435. PMID: 28285822.
Group Members
- Olivia Johnson
- Andrew Oliphant
- Andrew Patton
- Chee Sia
- Nicola Smyllie