

Replication can be arrested by DNA damage or by naturally occurring DNA secondary structures. We are particularly interested in translesion synthesis (TLS), which is mediated by specialised DNA polymerases and which is required for replication of both DNA damage and a particular secondary structure called a G quadruplex. Although TLS is potentially mutagenic it plays a critical role in normal cells and understanding its control is central to understanding cell transformation.
We study these processes using the powerful combination of vertebrate somatic cell genetics coupled with biochemical, biophysical and advanced optical microscopy techniques to monitor the molecular choreography of proteins and DNA at sites of stalled replication.
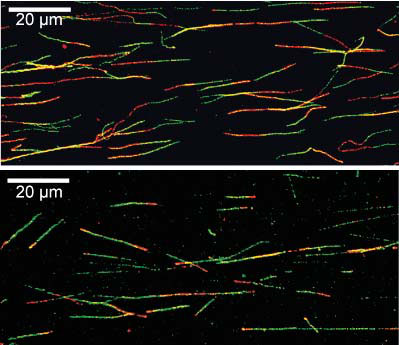
The green and red portions each reflect 20 minutes of DNA synthesis. DNA damage has been added
during the second (red) labelling period in the lower panel resulting in replication fork stalling.
Selected Papers
- Murat, P, Perez, C, Crisp, A, van Eijk, P, Reed, SH, Guilbaud, G, Sale, JE (2022)
DNA replication initiation shapes the mutational landscape and expression of the human genome
Science Advances 8(45): eadd3686. doi:10.1126/sciadv.add3686. - Mellor C, Perez, C, Sale, JE. (2022)
Creation and resolution of non-B-DNA structural impediments during replication
Crit Rev Biochem Mol Biol 57: 412-442 doi: 10.1080/10409238.2022.2121803 - Guilbaud G, Murat P, Wilkes HS, Lerner LK, Sale JE, Krude T. (2022)
Determination of human DNA replication origin position and efficiency reveals principles of initiation zone organisation
Nucleic Acids Res 50(13): 7436-7450. doi: 10.1093/nar/gkac555 - Murat P, Guilbaud G, Sale JE. (2020)
DNA polymerase stalling at structured DNA constrains the expansion of short tandem repeats.
Genome Biol. 21(1): 209. doi: 10.1186/s13059-020-02124-x. - Lerner LK, Holzer S, Kilkenny ML, Šviković S, Murat P, Schiavone D, Eldridge CB, Bittleston A, Maman JD, Branzei D, Stott K, Pellegrini L, Sale JE. (2020)
Timeless couples G-quadruplex detection with processing by DDX11 helicase during DNA replication.
EMBO J.e104185 doi: 10.15252/embj.2019104185. Online ahead of print. - Šviković, S., et. al. (2019)
R-loop formation during S phase is restricted by PrimPol-mediated repriming.
EMBO J. 38(3): pii: e99793, doi:10.15252/embj.201899793.
Group Members
- Patrick Cai
- Alastair Crisp
- Guillaume Guilbaud
- Emma Hands
- Julieta Molina Flores
- Joelle Nassar
- Sofia Obolenski-Koenig
- Harry Orrin
- Matthew Tayler
- Linda van Bijsterveldt
- Jan Verburg
- Andrew Zeller