

Gene transcription is an essential cellular process. Yet, RNA polymerase II (RNAPII) is constantly faced with challenges on its way across the gene. One of the main roadblocks to transcription are different types of DNA damage, which cause RNAPII stalling and prevent forward transcription. Several pathways have evolved to resolve stalled RNAPII (Figure 1, left) – we focus on understanding the mechanistic basis of these processes and relationships between them. Triggering these events on normally transcribing RNAPII complexes would be detrimental and incompatible with life. We investigate how cells direct these responses only at RNAPII molecules “in trouble”, at the correct time and place.
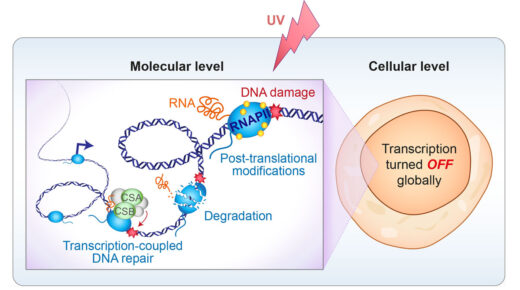
Transcription-blocking DNA lesions, such as those generated by UV-light, not only initiate mechanisms for resolving stalled RNAPII, but also induce a global shutdown of transcription genome-wide (Figure 1, right). Other types of cellular stress, even those that do not cause DNA damage, mimic several aspects of this global transcriptional response. Our lab aims to uncover how these processes operate at the molecular level.
Using a combination of genome-wide, biochemical and cell biology tools in mammalian systems (Figure 2), we study the fundamental aspects of the transcription process, in homeostasis and upon cellular stress such as DNA damage.
Selected Papers
- Cacioppo, R., Gillis, A., Shlamovitz, I., Zeller, A., Castiblanco, D., Crisp, A., Haworth, B., Arabiotorre, A., Abyaneh, P., Bao, Y., Sale, J.E., Berry, S., Tufegdžić Vidaković, A. (2024)
CRL3ARMC5 ubiquitin ligase and Integrator phosphatase form parallel mechanisms to control early stages of RNA Pol II transcription
Molecular Cell - Tufegdzic Vidakovic, A., Mitter, R., Kelly, G.P., Neumann, M., Harreman, M., Rodriguez-Martinez, M., Herlihy, A., Weems, J.C., Boeing, S., Encheva, V., et al. (2020)
Regulation of the RNAPII Pool Is Integral to the DNA Damage Response.
Cell 180: 1245-1261 e1221. - Tufegdzic Vidakovic, A., Harreman, M., Dirac-Svejstrup, A.B., Boeing, S., Roy, A., Encheva, V., Neumann, M., Wilson, M., Snijders, A.P., and Svejstrup, J.Q. (2019)
Analysis of RNA polymerase II ubiquitylation and proteasomal degradation.
Methods 159-160: 146-156.
Group Members
- Pegah Abyaneh
- Yu Bao
- Roberta Cacioppo
- Benjamin Haworth
- Pei Qin (Sabrina) Ng